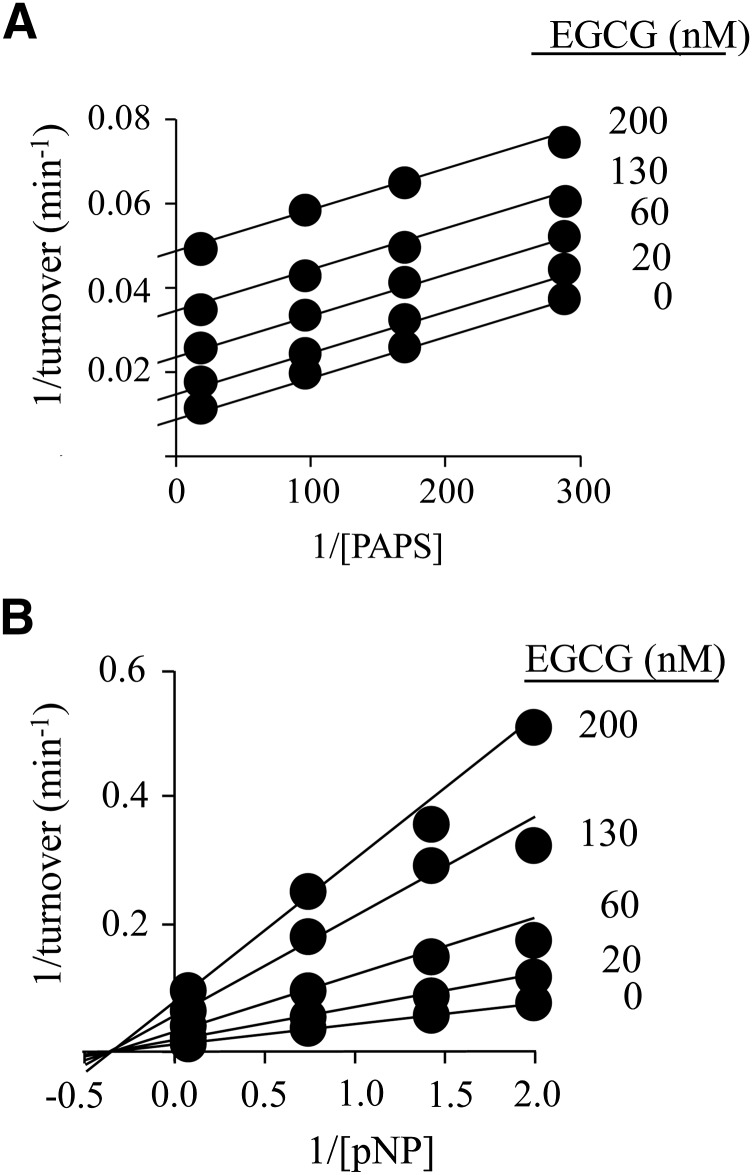
Fig. 1.
The inhibition of SULT1A1 by EGCG. (A) EGCG versus PAPS. PAPS concentration was varied from 0.2 to 5 × Km, and EGCG concentrations are given in the figure. Reactions were initiated by addition of pNP at saturation (30 μM, 20 × Km), and reaction progress was monitored by following formation of 35S-pNPS. Velocities were determined by least-squares fitting of four-point progress curves. Less than 5% of the concentration-limiting substrate consumed at the reaction endpoint was converted during the measurement. Velocities were determined in duplicate, averaged, and the data were fit globally using an uncompetitive model. The results of the fit are given by the solid lines passing through the data. (B) EGCG versus pNP. Reactions were initiated by addition of PAPS at saturation (10 μM, 625 × Km), the pNP concentration varied from 0.2 to 5 × Km, and the EGCG concentration is given in the figure. Reaction progress was monitored at 405 nm. Less than 5% of the concentration-limiting substrate consumed at the endpoint of the reaction was converted during the measurement. Each point represents the average of three independent determinations. The lines through the points represent the behavior predicted by a global fit using a noncompetitive inhibition model. Reaction conditions for both panels were as follows: SULT1A1 (10 nM, dimer), MgCl2 (5.0 mM), and NaPO4 (50 mM), pH = 7.2, and T = 25 ± °C.