Figure 4.
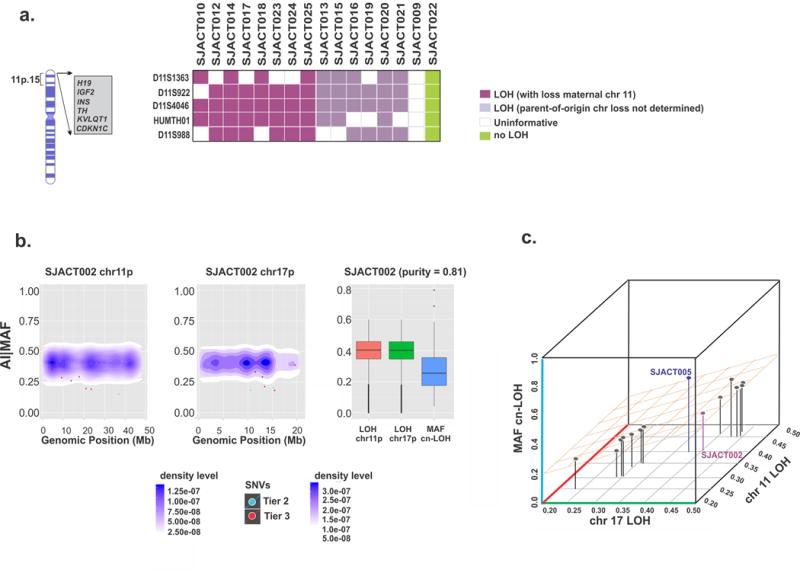
Characterization and timing of chromosome 11 and 17 LOH in pediatric ACT. (a) Microsatellite analysis of chromosome 11p15 in the WES cohort. All cases with available parental DNA demonstrated selective loss of maternal chromosome 11p15 (n=8, purple). (b) Temporal order of chromosome 11p and 17p cn-LOH and accumulation of single nucleotide variations (SNVs) in SJACT002. Scatter plots show mutant allele fractions (MAFs) of somatic SNVs and their genomic positions (individual dots) combined with 2-D density plots of allelic imbalance (AI) values of germline heterozygous SNPs in cn-LOH regions of chromosomes 11p (left) and 17p (center). At right, AI values in cn-LOH regions of chromosomes 11p and 17p were compared with the MAF distribution of somatic SNVs in genome-wide cn-LOH regions. (c) A 3-D scatter plot summarizes the temporal order of cn-LOH of chromosomes 11p and 17p and somatic SNV accumulation in pediatric ACTs. Shown are median AI values for the chromosome 11p cn-LOH region, median AI values for the chromosome 17p cn-LOH region and median MAF of SNVs in genome-wide cn-LOH regions of 14 cases; SJACT002 and SJACT005 are labeled. See also Supplementary Fig. 7b.
