Figure 5.
Post-Translational Modification by Arginine Methylation Alters the Phase Transition of Ddx4N1
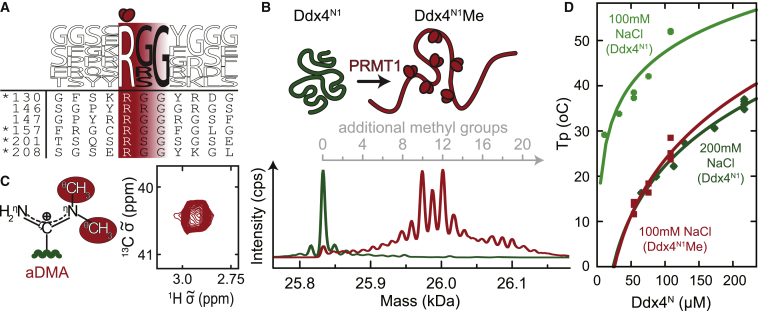
(A) Sequence logo (weblogo.berkeley.edu) depicting the amino acid motifs surrounding arginine residues of Ddx4N1 predominantly targeted by PRMT1. Arginine residues to be converted to aDMA are highlighted in dark red and with two small ellipses. The amino acid numbers of the modified arginine residues are shown within their respective sequence contexts. Asterisks highlight aDMA sites identified in Ddx4N1Me with 95% probability (Scaffold score) from a combination of trypsin and GluC digestion of recombinant, purified Ddx4N1Me. aDMA at sites 146 and 147 was identified at ∼65% probability (Scaffold score).
(B) Schematic and mass reconstruction of +TOF MS spectra of Ddx4N1 (green; 25.833 kDa) and Ddx4N1Me (dark red). In the latter, a series of peaks was observed between 1 and 20 methyl additions. The major peaks indicate complete aDMA modification at 5 and 6 sites, respectively.
(C) A schematic of aDMA together with an insert showing the 1H-13C HSQC NMR spectrum of the θCH3 of Ddx4N1Me. The chemical shifts of the methyl groups verify that the modification is aDMA (see Figure S5).
(D) The phase-transition temperatures of Ddx4N1Me (dark red) are shifted compared to the unmodified form under the same conditions (light green). Modification with aDMA at a mixture of 5–6 aDMA sites reduces the transition temperature by 25°C, an effect on the phase transition comparable to increasing the ionic strength by 100 mM.