Figure 1.
Crystal Structures UCH-L5/DEUBAD Complexes
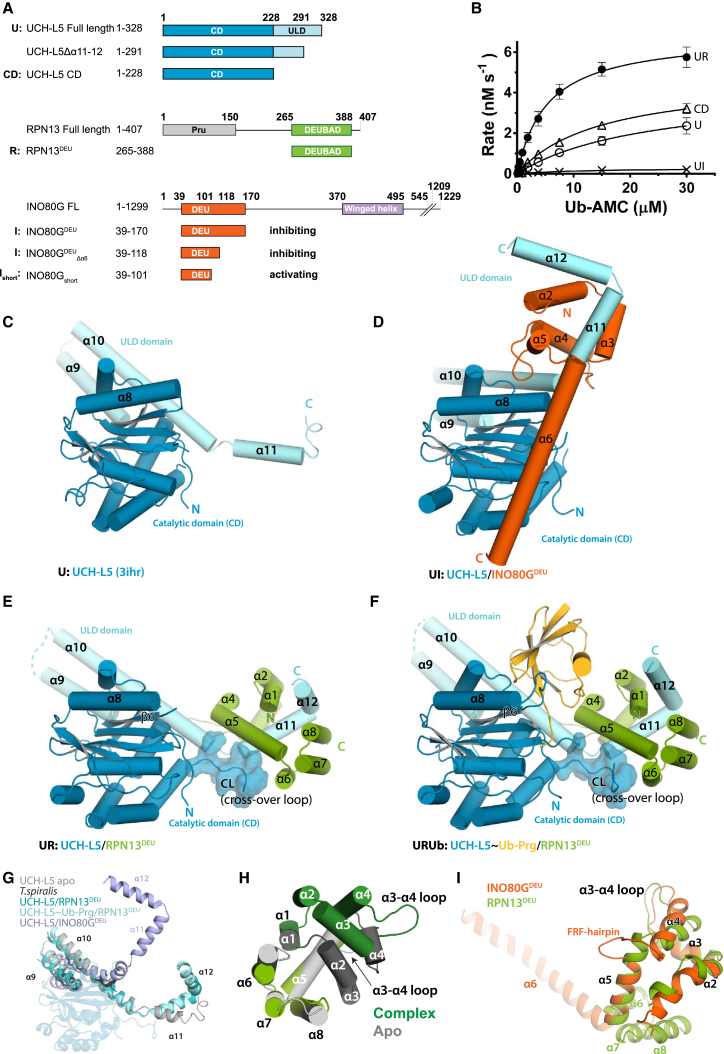
(A) Constructs used in this study.
(B) RPN13DEU activates UCH-L5 (UR) while INO80GDEU inhibits UCH-L5 (UI) in Ub-AMC enzyme kinetics. The CD is slightly more active than FL UCH-L5 (U). See Figure 1A for naming codes. Error bars, SD.
(C) Structure of apo UCH-L5 (3ihr). CD, blue; ULD domain, light blue.
(D) Structure of UCH-L5/INO80GDEU (INO80GDEU, orange).
(E) Structure of UCH-L5/RPN13DEU (RPN13DEU, green).
(F) Structure of UCH-L5∼Ub-Prg/RPN13DEU (Ub-Prg, yellow).
(G) The ULDs are found in different conformations across UCH-L5 structures. The CD is transparent for clarity.
(H) RPN13DEU (green) changes toward an open state upon UCH-L5 complex formation compared to apo RPN13DEU (gray). Helices α1–4 and the α3-4 loop that undergo the largest changes are colored in darker shades.
(I) Superposition of RPN13DEU and INO80GDEU. DEUBAD domains deviate most at FRF hairpin and helix α6. See also Tables 1, S1, S2, and S4 and Figure S1.