Ecologists recognize the balance of nature as a complex system that arises from interacting and connected parts; disturbing the balance can affect other species in nonlinear and dynamic ways. Like ecosystems, the cell and genome are extensively connected by an abundance of molecular interactions. Upsetting the molecular balance, through changes in gene regulation, abnormal copy number variants, and changes in cell size can create consequences for genome and organismal function (1, 2). In PNAS, Gibbons et al. (3) advance the idea of genome balance further by identifying a new source of genetic variation that results in balancing transcriptional subunits of the ribosome. Using data from human and mouse, the authors characterized copy number variation (CNV) in tandemly repeated ribosomal DNA (rDNA) arrays and found that subunits of the array vary together in what they define as concerted copy number variation (cCNV). cCNV raises new questions about how CNV can function to restore balance in eukaryotic genomes, especially on the assembly and function of the ribosome.
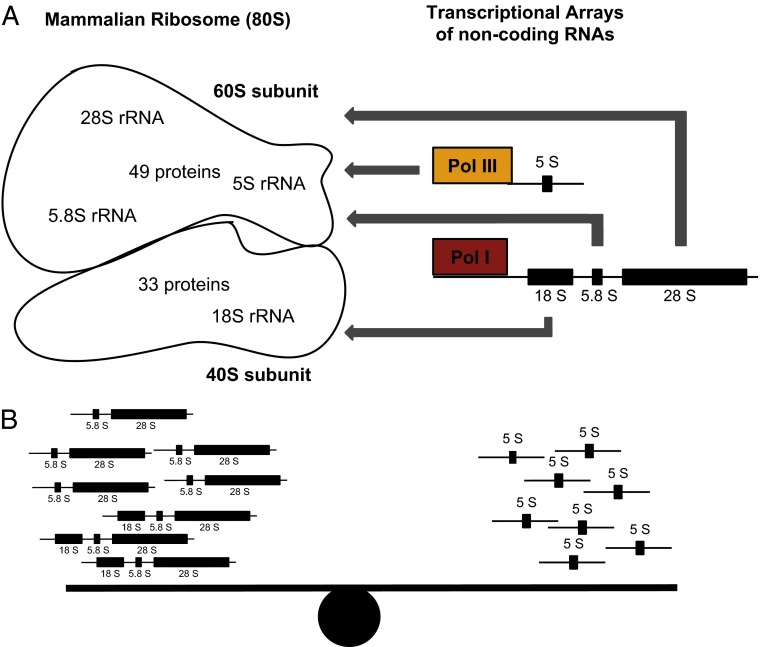
The eukaryotic ribosome is a macromolecular complex composed of about 80 single copy ribosomal proteins and 4 noncoding ribosomal RNAs whose genes are responsible for translating mRNAs into proteins in all living cells. The genes that encode these RNAs are extensively duplicated and scattered throughout the genome (4, 5). The noncoding RNA loci are especially intricate. The 45S rDNA locus is transcribed by RNA polymerase I and spliced to produce the 18S, 5.8S, and 28S rRNAs. The 5S rDNA locus is transcribed by polymerase III and produces 5S rRNA. These independently transcribed noncoding RNA units must act together to form the ribosome (Fig. 1A).
Fig. 1.
The mammalian ribosome is composed of a large subunit (60S) and a small subunit (40S) that are composed of a diverse set of proteins and transcriptional arrays of noncoding RNA loci (A). Gibbons et al. (3) discovered that CNV is coupled among transcriptional units at the DNA level, suggesting a new mechanism to achieve balance of the ribosome (B).
It seems challenging to coordinate the production of so many genes but there is indeed evidence for such coordination. How is the coordinated expression and production of so many proteins and RNAs to assemble the ribosome achieved? Common promoter motifs, hormone modulation of translation rates, duplicate copies and retrotransposition, tandem duplicate arrays, and feedback control loops are used in a variety of organisms to synchronize ribosome production (5). Therefore, there are diverse strategies to ensure an adequate supply of proteins and rRNAs. This flexibility is illustrated by the rapid changes in ribosome protein abundance that occurs in response to environmental signals, especially in response to changes in temperature, metabolism, and perturbation from chemicals (5–7). The ribosome then is a classic model of coordination and illustrates the diversity of ways to achieve balance, which if upset, results in detrimental consequences to the organism.
Although rapid responses of the level of ribosomal proteins are known, it is less clear how responses of ribosomal RNA gene clusters are expressed together. The tandem duplicated arrays of ribosomal RNA gene clusters account for over 80% of the RNA found in the cell, and these repeated arrays are some of the most evolutionarily dynamic regions of eukaryotic genomes, with extensive CNV found within and between species (4, 8). Because these arrays are unlinked, transcribed by different polymerases, and contribute to different parts of the ribosomes, it is interesting to wonder about the role of this extreme variation and how this affects ribosomal assembly.
Gibbons et al. (3) address this problem by using publically available human and mouse short-read DNA-Seq data and develop a computational method to measure copy number for each of the two independently transcribed arrays: the 45S rDNA locus and the 5S rDNA locus (Fig. 1A). The authors found more than 10-fold variation in DNA copy number, ranging from tens to hundreds of copies, for both the 45S and 5S rDNA loci and significant conservation of this variation between species. This conserved and extensive CNV reinforces the idea that rDNA loci are some of the most hypervariable regions of eukaryotic genomes.
Because the ribosome is composed of two main subunits, finding correlated variation in copy number raises the possibility that this concerted variation produces a balanced stoichiometry to preserve ribosome function. Probing this idea of balance further, Gibbons et al. (3) compared copy number assignments for each of the loci to determine the consistency. If CNV reflected the stoichiometric needs of the ribosome, then rDNA loci that contribute to the same subunits (the 5S, 5.8S, and 28S arrays) (Fig. 1A) should have more similar copies when compared with the 18S locus, which contributes to a different subunit. Consistent with this hypothesis, 5S, 5.8S, and 28S, all members of the large ribosomal subunit (60S), had the highest correlations in copy number compared with 18S, which contributes to the small ribosomal subunit (40S). These correlation analyses suggest that CNV is coordinated to match stoichiometric demands of each ribosomal subunit and may provide a way to achieve balance (Fig. 1B).
The intriguing observation that DNA copies are expanding and contracting concertedly in a sample of humans and mice raises questions about the speed at which these changes occur and how this variation may contribute to population variation. On the one hand, Gibbons et al. (3) find that the variation in copy number of rDNA is similar between humans and mouse, yet on the other hand, there is extensive intraspecific variation. To address how fast this cCNV variation is evolving, Gibbons et al. use pedigree analysis from several families to ask whether cCNV between the 5S and 45S loci covary from parents to offspring. Copy number expansion and contraction of the rDNA arrays varied across siblings and within families, supporting the idea that cCNV variation is rapidly evolving, and raising questions as to how this form of variation may contribute to population genetic structure.
To explore how these rapid amplifications and contractions of rDNA CNV relate to SNP variation, the authors compared SNP variation to CNV of the 5S rDNA array. CNV for 5S was nearly identical between populations (mean = 75.2 copies in Western Africa and 74.3 copies in Northern and Western Europe), whereas nearby SNP variation in the genome easily differentiates these populations. This finding suggests that cCNV variation is uncoupled from variation in nearby SNPs.
Finally, it has recently been shown that bisphenol A, an industrial chemical used in the making of plastics, disrupts expression of
Gibbons et al. advance the idea of genome balance further by identifying a new source of genetic variation that results in balancing transcriptional subunits of the ribosome.
genes coding for ribosomal proteins (7). Given the rapid cCNV in families, it is interesting to wonder how environmental perturbations like bisphenol A could affect the CNV of rDNA. Gibbons et al. (3) added bisphenol A to cells and measured the number of copies of rDNA loci relative to controls using quantitative PCR. Remarkably, the number of copies of rDNA loci was reduced compared with controls, and reduced in a way that paralleled cCNV. These experiments demonstrate that environmental triggers induce rapid and parallel loss in copies of 5S and 45S rDNA arrays and may provide mechanistic links between environmental perturbations and human health and disease.
The discovery that CNV can vary together, which is likely to balance the stoichiometry of the parts needed to assemble the ribosome, is a new source of genetic variation that expands the importance of CNV for understanding genome function, especially given the response of cCNV to environmental perturbations. It will be important to identify the mechanisms that produce these concerted copy numbers of rDNA loci, especially because the loci are unlinked, are spread throughout the genome, and have little sequence homology. How copy number is coordinated is not clear but perhaps some form of nonreplicative nonhomologous DNA repair contributes to this variation (9). Additionally, what happens if cCNV becomes uncoupled? Will this disrupt ribosome function? How do these copy number variants relate to expression levels of the rRNAs? Given the cCNV variation, presumably there will be a close relationship, but measuring the levels of RNA will help untangle the function of cCNV. With new genome-engineering technologies such as CRISPRs/cas (clustered regularly interspaced short palindromic repeats) and TALENs (transcription activator-like effector nucleases) (10), it may be possible to perturb CNV directly to explore how altering copy number among the rDNA subunits can affect balance of the ribosomal components and the consequences when changed. How does cCNV of rDNA vary with the single copy and expressed duplicates of the ribosomal proteins (11)? These new questions and the new areas of research that are inspired by the discovery of cCNV will be exciting to address as this field moves forward, but for now it appears that coordination among varying DNA copies is a new way to achieve gene and genome balance.
Footnotes
The author declares no conflict of interest.
See companion article on page 2485 in issue 8 of volume 112.
References
- 1.Birchler JA, Veitia RA. The gene balance hypothesis: Dosage effects in plants. Methods Mol Biol. 2014;1112:25–32. doi: 10.1007/978-1-62703-773-0_2. [DOI] [PubMed] [Google Scholar]
- 2.Birchler JA, Veitia RA. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA. 2012;109(37):14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibbons JG, Branco AT, Godinho SA, Yu S, Lemos B. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc Natl Acad Sci USA. 2015;112(8):2485–2490. doi: 10.1073/pnas.1416878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14(3):313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 5.Mager WH. Control of ribosomal protein gene expression. Biochim Biophys Acta. 1988;949(1):1–15. doi: 10.1016/0167-4781(88)90048-6. [DOI] [PubMed] [Google Scholar]
- 6.Xiao L, Grove A. Coordination of ribosomal protein and ribosomal RNA gene expression in response to TOR signaling. Curr Genomics. 2009;10(3):198–205. doi: 10.2174/138920209788185261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branco AT, Lemos B. High intake of dietary sugar enhances bisphenol A (BPA) disruption and reveals ribosome-mediated pathways of toxicity. Genetics. 2014;197(1):147–157. doi: 10.1534/genetics.114.163170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weider LJ, et al. The functional significance of ribosomal (r)DNA variation: Impacts on the evolutionary ecology of organisms. Annu Rev Ecol Evol Syst. 2005;36:219–242. [Google Scholar]
- 9.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10(8):551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng Y, et al. Making designer mutants in model organisms. Development. 2014;141(21):4042–4054. doi: 10.1242/dev.102186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dharia AP, Obla A, Gajdosik MD, Simon A, Nelson CE. Tempo and mode of gene duplication in mammalian ribosomal protein evolution. PLoS ONE. 2014;9(11):e111721. doi: 10.1371/journal.pone.0111721. [DOI] [PMC free article] [PubMed] [Google Scholar]