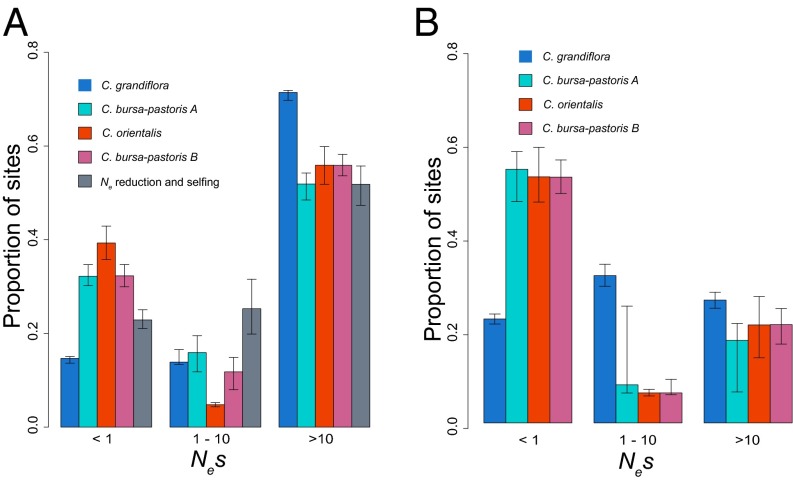
Fig. 4.
Estimates of the DFE of deleterious mutations at zerofold nonsynonymous sites (A) and conserved noncoding sites (B), based upon the allele frequency spectra of SNPs at these sites. The simulated DFE for zerofold nonsynonymous sites under Ne reduction and a shift to selfing is also shown in A. The strength of selection is measured as Nes, where s is the strength of selection. Error bars correspond to 95% confidence intervals of 200 bootstrap replicates of 10-kb blocks.