Abstract
Genomic imprinting is an epigenetic mechanism that restrains the expression of ∼100 eutherian genes in a parent-of-origin-specific manner. The reason for this selective targeting of genes with seemingly disparate molecular functions is unclear. In the present work, we show that imprinted genes are coexpressed in a network that is regulated at the transition from proliferation to quiescence and differentiation during fibroblast cell cycle withdrawal, adipogenesis in vitro, and muscle regeneration in vivo. Imprinted gene regulation is not linked to alteration of DNA methylation or to perturbation of monoallelic, parent-of-origin-dependent expression. Overexpression and knockdown of imprinted gene expression alters the sensitivity of preadipocytes to contact inhibition and adipogenic differentiation. In silico and in cellulo experiments showed that the imprinted gene network includes biallelically expressed, nonimprinted genes. These control the extracellular matrix composition, cell adhesion, cell junction, and extracellular matrix-activated and growth factor–activated signaling. These observations show that imprinted genes share a common biological process that may account for their seemingly diverse roles in embryonic development, obesity, diabetes, muscle physiology, and neoplasm.
Genomic imprinting is an epigenetic phenomenon that leads to a parent-of-origin-specific expression of alleles in metatherian and eutherian mammals (Ferguson-Smith 2011). Since its discovery in the mid-1980s (McGrath and Solter 1984; Surani et al. 1984; Cattanach and Kirk 1985), it has been the focus of intense investigations; yet the reason for its implementation during evolution is still debated (Renfree et al. 2009). A major focus in genomic imprinting is the mechanisms underlying parental origin–specific expression of imprinted genes (IGs). The type of epigenetic marks, how they are established at specific loci during gametogenesis, and how they are maintained after fertilization, as well as the mechanisms leading to monoallelic expression, are now understood for several IGs (Bartolomei and Ferguson-Smith 2011; Kelsey and Feil 2013). However, a global analysis of IGs to discover a possible common role in cell function has not been comprehensively examined so far.
Pronuclear transplantation experiments (McGrath and Solter 1984; Surani et al. 1984), the analysis of the phenotype of mouse strains carrying uniparental disomy of various chromosomes or uniparental duplication of various chromosomal regions (Cattanach and Kirk 1985; Cattanach et al. 2006), and the phenotypic characterization of targeted mouse mutants led to the suggestion that IGs are key regulators of embryonic development (Ferguson-Smith 2011). The role of IGs is not limited to embryonic development, however, because alteration of imprinted loci leads to various pathological conditions in adult humans, including obesity, diabetes, muscle hypertrophy, mental disability, and neoplasm. Molecular functions associated with IGs are diverse and include signaling, ion channel, nutrient transport, transcription factor, noncoding RNA, extracellular matrix (ECM) protein, control of cell cycle, metabolism, protein synthesis, protein degradation, and vesicular secretion. Apart from Ins2, Igf2, Igf2r, and Grb10, which are members of the insulin/insulin-like growth factor signaling pathway, the molecular functions of IGs seem unrelated.
We previously demonstrated that a subset of 15 IGs is frequently coexpressed and likely belongs to the same gene network (Varrault et al. 2006). The present work extended this discovery and showed that most IGs belonged to a single gene network that also comprised biallelically expressed genes involved in the control of the ECM composition, cell adhesion, and cell–cell contacts. Furthermore, we provided evidence that the core function associated with this network was the control of cell cycle withdrawal/reentry and differentiation, possibly through ECM recomposition.
Results
Coexpression of IGs
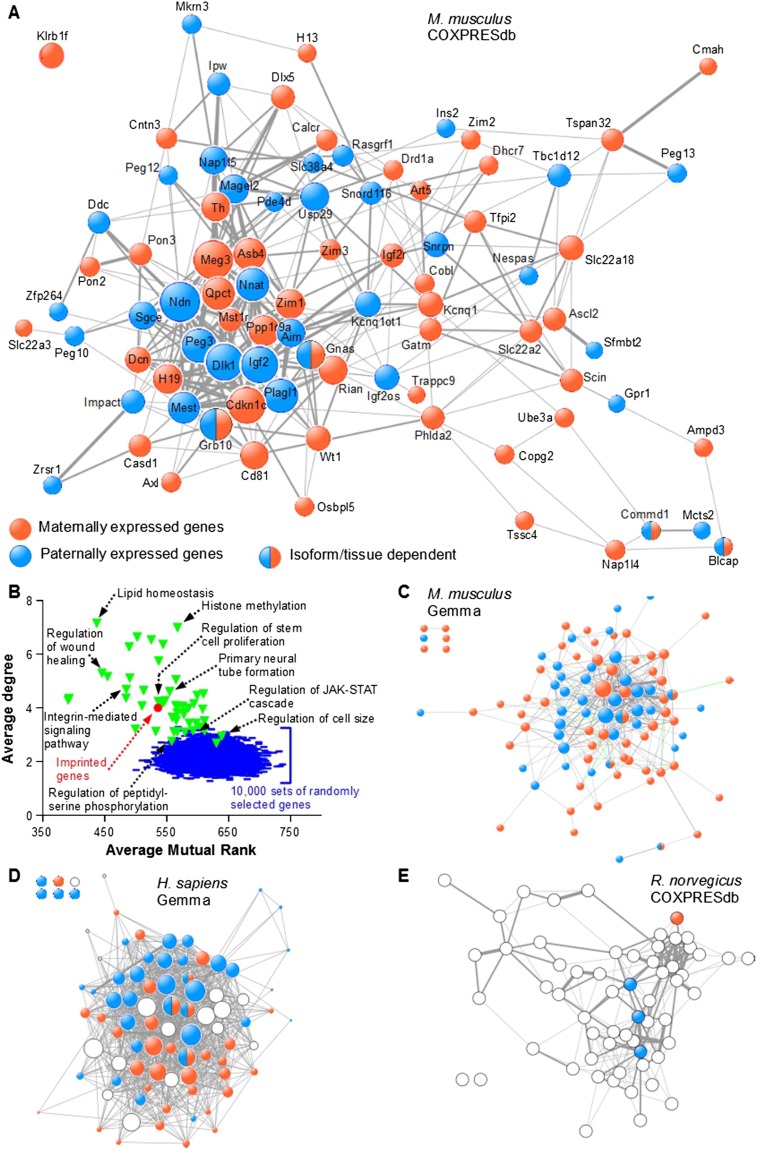
We performed functional annotation and classification of murine IGs (Supplemental Table S1) using the DAVID bioinformatics resources (http://david.abcc.ncifcrf.gov/) and found no enrichment of specific Gene Ontology terms and KEGG pathways (Supplemental Table S2), which did not support a common function for IGs. However, using a meta-analysis of microarray data (Lee et al. 2004a), we previously showed that 15 IGs are frequently coregulated (Varrault et al. 2006). In the present study, we extended this discovery to all IGs by mining two larger meta-analyses, COXPRESdb (Obayashi and Kinoshita 2011) and Gemma (Zoubarev et al. 2012), each one using a distinct metric and relying on diverse data sets. We first searched COXPRESdb for coexpression of the 85 murine IGs present in this database, and showed that 84 are frequently coexpressed (Fig. 1A; Supplemental Table S3). Details regarding the construction of this network are given in the Supplemental Information. To test for the specificity of this network, we compared its topology, i.e., the average number of neighbors of a node (degree) and the average edge weight (mutual rank in COXPRESdb), to coexpression networks made from other gene sets. We showed that the network of 85 IGs was topologically different to 10,000 networks obtained by randomly drawing sets of 85 genes with data in COXPRESdb (Fig. 1B). We also constructed 50 coexpression networks from sets of 85 genes known to be functionally related, i.e., belonging to the same Gene Ontology Biological Process (GO_BP) (Fig. 1B; for the full list of GO_BPs used in this figure, see Supplemental Table S22; for details regarding the selection of GO_BPs, see Supplemental Information). A small number of these networks, e.g., the network made from genes that belong to the GO_BP “Regulation of peptidyl-serine phosphorylation,” were topologically similar to random networks (Fig. 1B), indicating that these genes were not transcriptionally coregulated. However, most networks made from genes in the same GO_BP were different to random networks. Interestingly, the network of IGs was similar to networks made from genes belonging to the same GO_BP. To verify that coexpression of IGs was not an artifact of the metric and/or data sets used in COXPRESdb, we performed a similar analysis with the 101 murine IGs present in Gemma and showed that 95 were frequently coexpressed (Fig. 1C; Supplemental Table S4). As we did previously with the COXPRESdb network (Fig. 1B), we verified that the Gemma network of murine IGs is topologically similar to coexpression networks made from GO_BP genes and different to networks made from randomly selected genes (Supplemental Fig. S1). We compared the COXPRESdb and Gemma networks to figure out how similar they are. We first looked at the overall topology of both networks and measured the degree correlation (Supplemental Fig. S2). The degree of each IG in both of the databases is well correlated (r2 = 0.70), indicating that genes highly connected in one database were generally highly connected in the other. We then recorded edges between the 80 murine IGs jointly found in the networks displayed in Figure 1A and C, and classified them in three categories (Supplemental Fig. S3): present in both databases (139 edges), present only in Gemma (176 edges), and present only in COXPRESdb (181 edges). This classification qualitatively showed that connections among a small group of highly connected IGs are well conserved in COXPRESdb and Gemma. We also plotted the relative difference of degree in the two databases versus the average degree of each gene in the two databases (Supplemental Fig. S4). This quantitative analysis suggested that three groups of coexpressed IGs existed. A group of 13 IGs—i.e., Meg3, Ndn, Grb10, Dlk1, Igf2, Cdkn1c, Plagl1, Peg3, Mest, Nnat, Asb4, H19, and Ppp1r9a—were highly connected with other IGs (average degree ≥15), and both the number and the identity of these connections were conserved in Gemma and COXPRESdb (relative degree difference ≤50%). This subnetwork was apparent in Figure 1A as the densely connected nodes in the lower left quadrant of the figure. A second group of 10 IGs included Gnas, Magel2, Rian, Dcn, Snrpn, Airn, Cd81, Igf2os, Impact, Phlda2, and Gatm. These genes displayed intermediate connectivity, and their connections were less frequently conserved in the two databases. Finally, the other IGs displayed less connections in at least one database (low average degree) and/or a large relative difference of degree in the two databases.
Figure 1.
Imprinted genes are frequently coexpressed. (A) The COXPRESdb meta-analysis of microarray data was searched for coexpression among murine IGs. The resulting coexpression links are represented using Cytoscape. Node size is proportional to node degree. Edge width represents the mutual rank (see Supplemental Information) between two given nodes. (B) The average degree and average mutual rank were computed for the network of 85 IGs displayed in A (red circle). Random networks (blue lines) were generated by randomly drawing 10,000 sets of 85 Gene IDs with data in COXPRESdb and retrieving coexpression links as for the 85 murine IGs. A similar procedure was performed with 50 sets of 85 genes found in GO Biological Processes (green triangles) and with data in COXPRESdb (for a full list of the GO BPs analyzed, see Supplemental Table S22). (C) The Gemma meta-analysis of microarray data was searched for coexpression among murine IGs. Node size is proportional to node degree. Edge width maps the number of data sets in which two given nodes are coexpressed. (D) The Gemma database was searched for coexpression among human IGs and orthologs of murine IGs whose imprinting status in humans is unknown or uncertain (open circles) as in C. (E) The COXPRESdb was searched for coexpression of rat IGs and orthologs of murine IGs whose imprinting status in rat is unknown (open circles) as in A.
We performed a similar analysis with the 86 human IGs (Supplemental Table S5) present in Gemma, including 15 genes whose imprinting status is unknown but whose murine orthologs are imprinted. Again, the vast majority of IGs (80 out of 86) were frequently coexpressed (Fig. 1D; Supplemental Table S6). We verified that the network of human IGs is topologically similar to coexpression networks made from GO_BP genes and different to networks made from randomly selected genes (Supplemental Fig. S5). Finally, we looked at rat IG coexpression and included in the analysis the rat orthologs of murine IGs (Supplemental Table S7). We showed that 58 out of 60 rat bona fide and candidate IGs were frequently coexpressed (Fig. 1E; Supplemental Table S8). Collectively, our data showed that IGs were coexpressed irrespective of the coexpression metric, data sets, and species, supporting our premise that they work in the same gene network.
IGs were coregulated during cell cycle withdrawal/reentry and differentiation
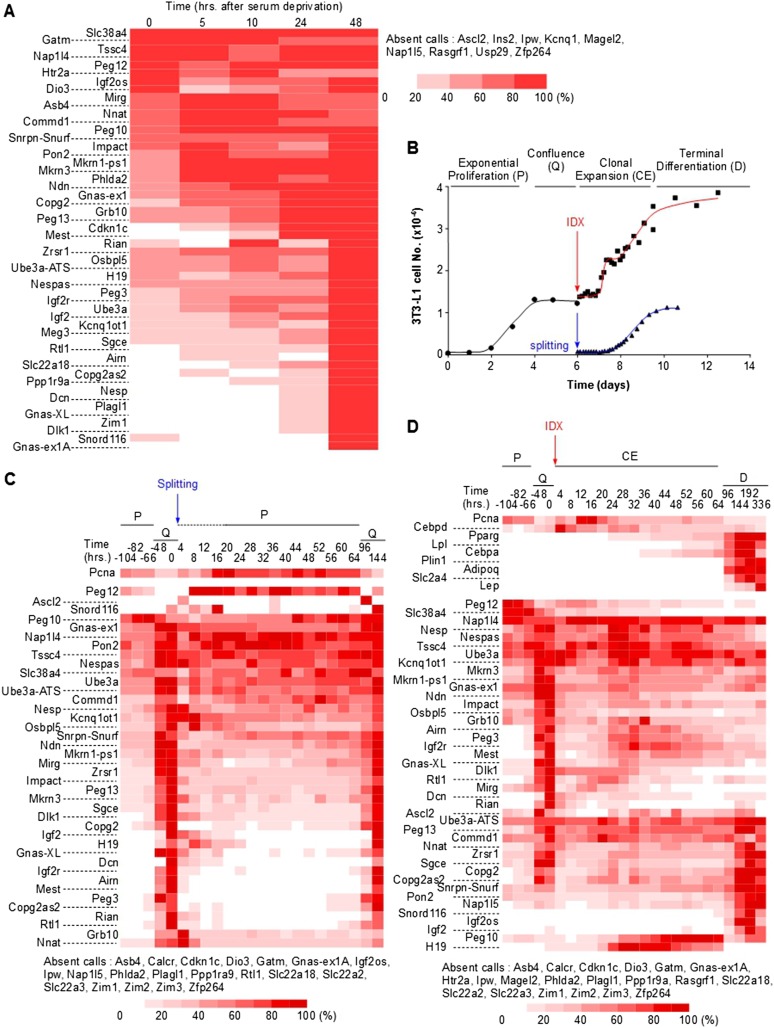
To gain insights into the biological function associated with this network, we searched for biological conditions under which IGs are dynamically coregulated. We first examined the data sets that displayed coexpression of many IGs and noticed that some were from cell populations whose proliferation was experimentally manipulated (data not shown). Using a collection of primers for real-time PCR, we monitored IG expression in primary mouse embryonic fibroblasts. We first induced cell cycle withdrawal by growth factor deprivation and observed the up-regulation of a large proportion of the tested IGs (Fig. 2A). We also let the cells reach confluence in the presence of serum to induce quiescence by contact inhibition, and observed a marked induction of the expression levels of most IGs upon MEF cell cycle exit (Supplemental Fig. S6). Down-regulation of IGs following splitting of confluent MEFs occurred as early as 4 h after splitting (Supplemental Fig. S6), far before the cells reenter the S phase ∼16 h after splitting (data not shown). This suggested that IG down-regulation was an early event when cells took the decision to leave G0 to reenter the cell cycle.
Figure 2.
Imprinted genes are coregulated upon cell cycle exit/reentry and differentiation. (A) Heatmap of IG expression levels following serum withdrawal. Exponentially growing mouse embryonic fibroblasts were serum-starved, and expression levels of the indicated IGs were monitored by real-time PCR at the indicated time after serum withdrawal. Data for each IG are expressed as the percentage of the maximal expression levels for that IG. (B) The 3T3-L1 adipogenic differentiation model. 3T3-L1 preadipocytes were grown exponentially (P, proliferation) until they reached confluence (Q, quiescence) following contact inhibition. Forty-eight hours later, they were either split and resumed exponential growth, or induced to differentiate following addition of IDX. In the latter condition, induced preadipocytes resumed proliferation during the clonal expansion phase (CE, clonal expansion), and eventually exited the cell cycle and differentiated (D, differentiation). (C) Heatmap of IG expression levels during 3T3-L1 exponential growth and quiescence. 3T3-L1 preadipocytes were grown as described in B. Expression levels of the indicated IGs were monitored and depicted as in A. Pcna is a marker of cell proliferation. (D) Heatmap of IG expression levels during 3T3-L1 adipogenic differentiation. 3T3-L1 preadipocytes were grown as in B. Expression levels of the indicated IGs were monitored and represented as in A. Cebpd, Pparg, Lpl, Cebpa, Plin1, Adipoq, Slc2a4, and Lep are markers of early and late adipogenic differentiation.
We then used another cell system, the 3T3-L1 preadipocyte cell line (Green and Kehinde 1974), in which we could finely tune the proliferation and differentiation status by splitting the cells or adding IDX (insulin, dexamethasone, and isobutylmethylxanthine) (Fig. 2B). As in MEFs, we observed that most IGs were minimally expressed during 3T3-L1 exponential growth and induced when cells reached confluence following contact inhibition (Fig. 2C). When quiescent 3T3-L1 cells were incubated with serum and IDX, the cells retracted and reentered the cell cycle in the so-called clonal expansion phase. The resulting synchronous proliferation took place in a different context, however, as the cells received the whole information (IDX) to undergo adipogenic differentiation. Interestingly, we again observed a rapid down-regulation of most IGs upon IDX addition (Fig. 2D). Two to three days following IDX addition, the cells definitively exited the cell cycle (Fig. 2B,D). It was noteworthy that only a subset of IGs was then reinduced, indicating a differential IG expression pattern depending on the reversibility of the cell cycle withdrawal. Finally, Peg10 and H19 displayed a specific pattern of expression with maximal expression during the second cell cycle that preceded definitive cell cycle exit (Fig. 2D).
To verify that our observations were also valid in vivo, we looked at a more physiological system in which a relatively homogenous cell population undergoes proliferation and differentiation in a controlled fashion. We studied muscle regeneration following injection of notexin, a snake venom phospholipase A2 that inhibits neuromuscular transmission by blocking acetylcholine release and is directly toxic to skeletal muscle upon local application in vivo. We injected notexin into the tibialis anterior muscle of adult mice and collected muscles at different time points (Supplemental Fig. S7). Within 2 d following notexin injection, the muscle is infiltrated with numerous macrophages that clear the muscle from degenerating myocytes. In parallel, satellite cells—the muscle stem cells—massively reenter proliferation to generate large numbers of myoblasts that further proliferate, exit the cell cycle, and ultimately differentiate into new myocytes. This in vivo regeneration system obviously does not enable the same control over the transition from proliferation to cell cycle exit and differentiation as the adipogenic differentiation paradigm previously studied. Yet, the pattern of expression of proliferation, inflammation, and differentiation markers allowed identifying the main period of cell cycle exit and initiation of differentiation. Proliferation and inflammation were most prominent until day 3 (Supplemental Fig. S7). From day 3 onward, the myogenic differentiation program gradually took place until day 15, when the pattern of gene expression closely resembled that of control muscle. Remarkably, IGs were induced in sequential waves, mainly between days 3 and 6, when the myoblasts stopped proliferating and initiated the myogenic differentiation process (Supplemental Fig. S7). These in vivo data corroborated our previous data showing prominent induction of IGs at the transition from proliferation to cell cycle withdrawal/differentiation.
IG regulation did not involve alteration of DNA methylation and biallelic expression
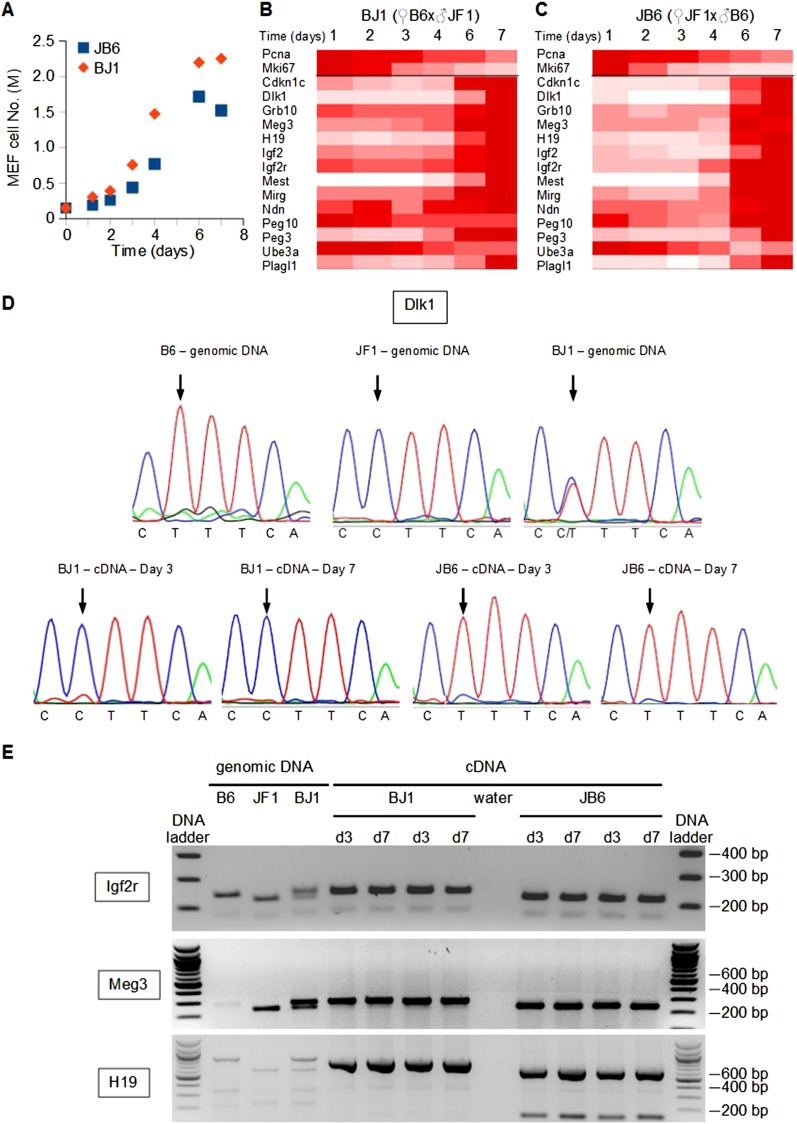
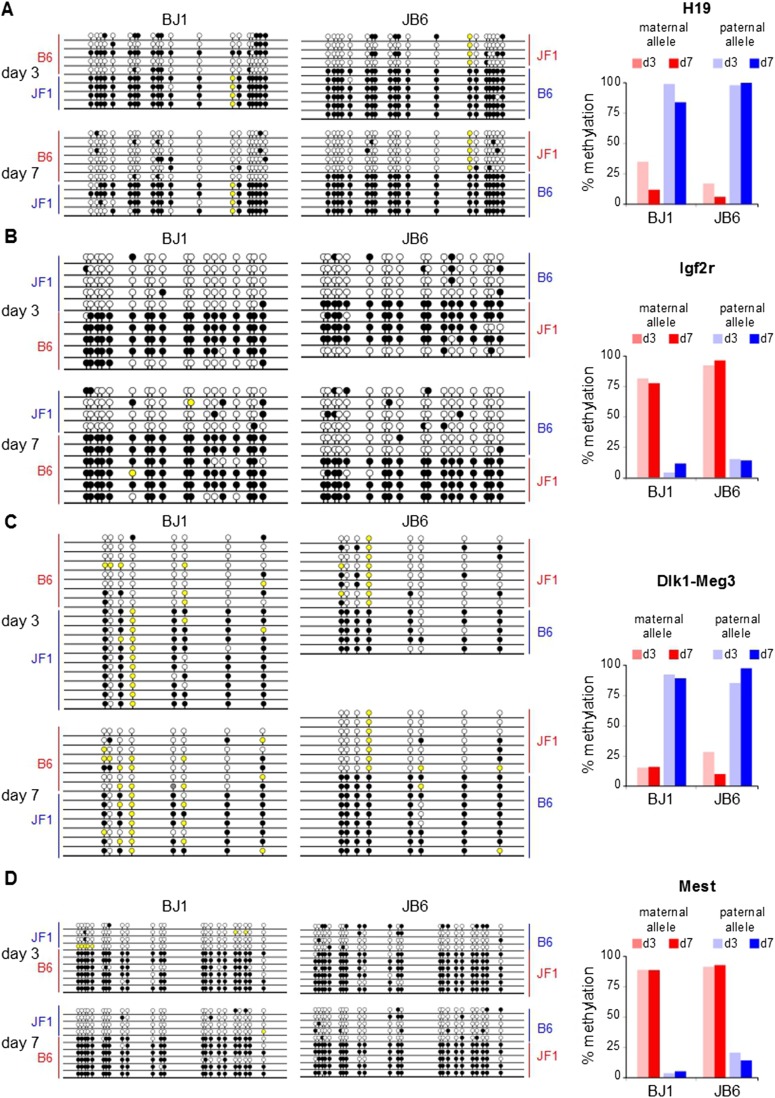
Because IGs are monoallelically expressed, the IG up-regulation we observed might result from the alteration of genomic imprinting and the subsequent derepression of the silenced allele. When MEFs from reciprocal crosses between C57BL/6J and JF1/Ms strains were grown in vitro until confluence (Fig. 3A), we observed the expected up-regulation of several IGs in quiescent MEFs (Fig. 3B,C). From BJ1 and JB6 MEF cDNAs, we PCR amplified a portion of the Dlk1 transcript that includes a single-nucleotide polymorphism to distinguish the C57BL/6J and JF1/Ms alleles. Sanger sequencing of the resulting amplicons demonstrated that only the paternal allele was expressed, irrespective of the proliferation status of the cells (Fig. 3D). Using polymorphisms that result in different amplicon size (Igf2r) or restriction fragment length polymorphism (RFLP) (Meg3 and H19), we showed that Igf2r, Meg3, and H19 were monoallelically expressed irrespective of the proliferation status of the cells (Fig. 3E). We then tested whether the proliferation status of MEFs influenced the DNA methylation pattern at imprinted loci. We treated genomic DNA from proliferating (day 3) and quiescent (day 7) BJ1 and JB6 MEFs with sodium bisulfite, which transforms cytosine to uracil and leaves methylcytosine unchanged. Sequencing of amplicons from bisulfite-treated genomic DNA revealed that the methylation patterns of the paternal and maternal alleles of the H19 (Fig. 4A), Igf2r (Fig. 4B), Dlk1-Meg3 (Fig. 4C), and Mest (Fig. 4D) loci did not change with the proliferation status of the MEFs. We also monitored the methylation status of the Meg3 promoter by the same technique and found no evidence of promoter methylation alteration upon cell cycle exit (Supplemental Fig. S8A). We verified our findings using another technique based on the digestion of genomic DNA by the methylation-dependent enzyme McrBC followed by PCR amplification. Again, we did not observe a significant alteration of the methylation status of several imprinted loci, including Peg3, Kcnq1ot1, H19-Igf2, Igf2r, and Dlk1-Meg3 (Supplemental Fig. S8B). All in all, our data demonstrated that the IG up-regulation we observed upon cell cycle withdrawal was not attributable to an alteration of the methylation pattern at imprinted loci or to a switch from mono- to biallelic expression.
Figure 3.
Monoallelic, parent-of-origin-dependent expression of imprinted genes is not altered in quiescent versus proliferating mouse embryonic fibroblasts. (A) Time course of cell numbers. JB6 and BJ1 MEFs were grown in vitro until they reached confluence. (B,C) Expression levels of proliferation markers (Pcna, Mki67) and representative IGs were monitored by real-time PCR in JB6 (B) and BJ1 (C) MEFs. Data for each gene are expressed as the percentage of the maximal expression levels for that gene. (D) Sequence of a polymorphic region (C[T/C]TTCA) of the Dlk1 gene (top panels) and transcripts (bottom panels). Genomic DNAs from C57BL/6J (B6), JF1/Ms, and C57BL/6J × JF1/Ms (BJ1) were sequenced and display the T, C, and C/T alleles, respectively. Sequencing of cDNAs from proliferating (day 3) and quiescent (day 7) MEFs derived from C57BL/6J × JF1/Ms (BJ1) and JF1/Ms × C57BL/6J (JB6) crosses indicates that only the paternal Dlk1 allele is expressed. (E) Genomic DNA and cDNAs were PCR amplified at the indicated loci from the indicated crosses. The amplicons from the Meg3 and H19 loci were digested with Bsh1236I and BclI, respectively. Digested and undigested amplicons were run on an ethidium bromide–stained agarose gel.
Figure 4.
Methylation pattern of differentially methylated regions at IG loci. Bisulfite-treated genomic DNAs from BJ1 and JB6 MEFs grown in vitro for the indicated period of time were PCR amplified at the indicated loci (A, H19; B, Igf2r; C, Dlk1-Meg3; D, Mest), subcloned, and sequenced. Filled and open circles denote methylated and unmethylated CpGs, respectively. Yellow circles denote variation from the corresponding C57BL/6J reference genome. B6 and JF1 indicate the parental allele from which each cloned amplicon is derived. Panels on the right side of the figure display the percentage of methylated maternal or paternal allele at each locus and each time point.
Alteration of IG expression altered cell cycle exit and adipogenic differentiation
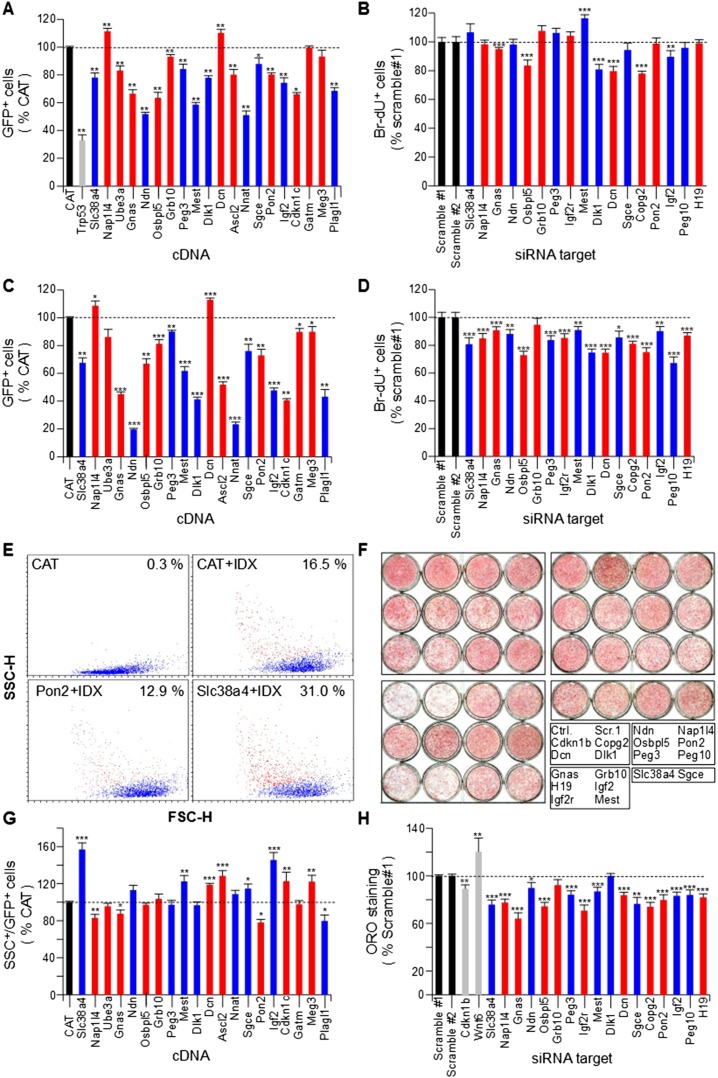
The experiments depicted in Figure 2 and Supplemental Figures S6 and S7 were suggestive of a role for IGs in the transition from proliferation to quiescence/differentiation in various cellular systems. We performed gain and loss of IG function to test whether alteration of IG expression did result in altered proliferation and/or differentiation. We selected several IGs based on their expression pattern in the adipogenic differentiation model and transiently transfected 3T3-L1 cells with siRNAs or cDNAs for 17 and 20 IGs, respectively. SiRNAs were carefully selected to potently induce >70% knockdown of the expression levels of the target gene in confluent 3T3-L1 cells (Supplemental Fig. S9). The amount of transfected cDNA resulted in low to moderate overexpression when compared to confluent 3T3-L1 and/or MEFs (Supplemental Table S9). We verified that cDNA overexpression was not overly toxic and measured apoptosis following cDNA transfection (Supplemental Fig. S10). Transfection of most IG cDNAs did not result in apoptosis of the transfected cells compared to CAT-transfected cells. Plagl1 was as potent as Trp53, as was expected from our published data (Spengler et al. 1997), and Gnas, Osbpl5, Grb10, Ascl2, and Cdkn1c induced apoptosis to a limited and variable extent compared to Trp53 and Plagl1. Overexpression of most IGs reduced the proliferation rate, except for Nap1l4 and Dcn (Fig. 5A). In contrast, down-regulation of IG expression in exponentially growing cells only had a weak effect on the proliferation rate (Fig. 5B), confirming that the minimal IG expression levels observed during exponential growth correspond to a mostly inactive state of the IG network with respect to proliferation. Exogenous expression of IGs while the endogenous IGs were down-regulated at the inception of the clonal expansion phase mostly resulted in reduced proliferation rates, except for Nap1l4 and Dcn (Fig. 5C), which confirmed that sustained IG expression tends to restrain proliferation. When IGs were down-regulated just before cell cycle exit (Fig. 5D), the effect on the proliferation rate was more pronounced than during exponential growth (Fig. 5B), confirming that the increased expression levels of IGs during quiescence entry were functionally relevant.
Figure 5.
Modulation of imprinted gene expression alters cell cycle exit and adipogenic differentiation of 3T3-L1 preadipocytes. (A) Effect of IG overexpression on cell numbers during exponential growth. Exponentially growing 3T3-L1 cells were cotransfected with eGFP and CAT (chloramphenicol acetyl transferase) as a negative control, Trp53 as a positive control of cell growth inhibition, or cDNAs encoding the indicated IGs. Following plating at low density, GFP-positive cells were counted 48 h post-transfection (n = 8): 84.8 ± 6.4% CAT-transfected cells were GFP positive. (B) Effect of IG down-regulation on DNA synthesis during exponential growth. Exponentially growing 3T3-L1 cells were transfected with siRNAs targeting the indicated IGs and plated at low density. DNA synthesis was monitored by measuring BrdU incorporation 24 h post-transfection (n = 20) and compared to the appropriate control condition, which displayed 31.9 ± 4.0% BrdU-positive cells. (C) Effect of IG overexpression on cell numbers during clonal expansion. 3T3-L1 cells were cotransfected as in A and plated at confluence. Three days post-transfection, cells were incubated with IDX to trigger adipogenic differentiation. GFP-positive cells were counted 8 d after IDX addition (n = 14): 26.2 ± 5.2% CAT-transfected cells were GFP positive. (D) Effect of IG down-regulation on DNA synthesis when cells reach confluence. 3T3-L1 cells were transfected as in B and plated at mid-confluence. DNA synthesis was monitored as in B (n = 20 for Ndn and Sgce; n = 30 for all other IGs): 11.5 ± 2.4% of control cells were BrdU positive. (E) Effect of Pon2 and Slc38a4 overexpression on adipogenic differentiation of 3T3-L1 preadipocytes. Cells were transfected as in A and plated at confluence. Eight days after IDX addition, cell size (forward light scattering [FSC]) and granularity (side light scattering [SSC]) of GFP-positive cells were measured by flow cytometry. SSC allows visualizing accumulation of lipid vesicles in differentiated adipocytes (red dots). (F) Effect of IG down-regulation on adipogenic differentiation of 3T3-L1 preadipocytes. Representative Oil Red O (ORO, a lipid stain) staining of 3T3-L1 cells that were left untransfected (Ctrl.) or transfected with scramble siRNA (Scr.1) or with siRNAs targeting the indicated genes, plated in duplicate at confluence, incubated with IDX 3 d post-transfection, and fixed 12 d later (n = 20 for Ndn and Sgce; n = 30 for all other IGs). (G) Effect of IG overexpression on adipogenic differentiation of 3T3-L1 preadipocytes. Cells were transfected and treated as in A. Quantification of SSC-, GFP-positive cells after transfection and induction of differentiation, compared to the CAT negative control at day 8 of differentiation (n = 16). (H) Effect of IG down-regulation on adipogenic differentiation of 3T3-L1 preadipocytes. Cells treated as in B were stained with ORO, which was quantified as described in Methods (n = 30). Cdkn1b and Wnt6 are positive controls negatively and positively affecting the adipogenic differentiation process, respectively. Data are mean ± SEM of the indicated number of replicate measures from at least three independent experiments. Statistical significance was assessed using a nonparametric, pairwise Wilcoxon test. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
To test the effect of the gain of IG function on adipogenic differentiation, we transfected preadipocytes to induce sustained IG expression, added IDX 48 h after confluence, and monitored adipogenic differentiation 10–14 d later. We monitored lipid vesicle formation in cDNA-transfected, GFP-positive cells by measuring light side scattering (SSC) (Fig. 5E; Supplemental Fig. S11). Similarly, lipid accumulation in siRNA-transfected cells was monitored by Oil red O (ORO) staining (Fig. 5F). Interestingly, gain and loss of most IG expression significantly altered lipid accumulation in 3T3-L1 adipocytes (Fig. 5G,H).
The IGN comprised biallelically expressed genes involved in ECM remodeling and ECM-linked signaling
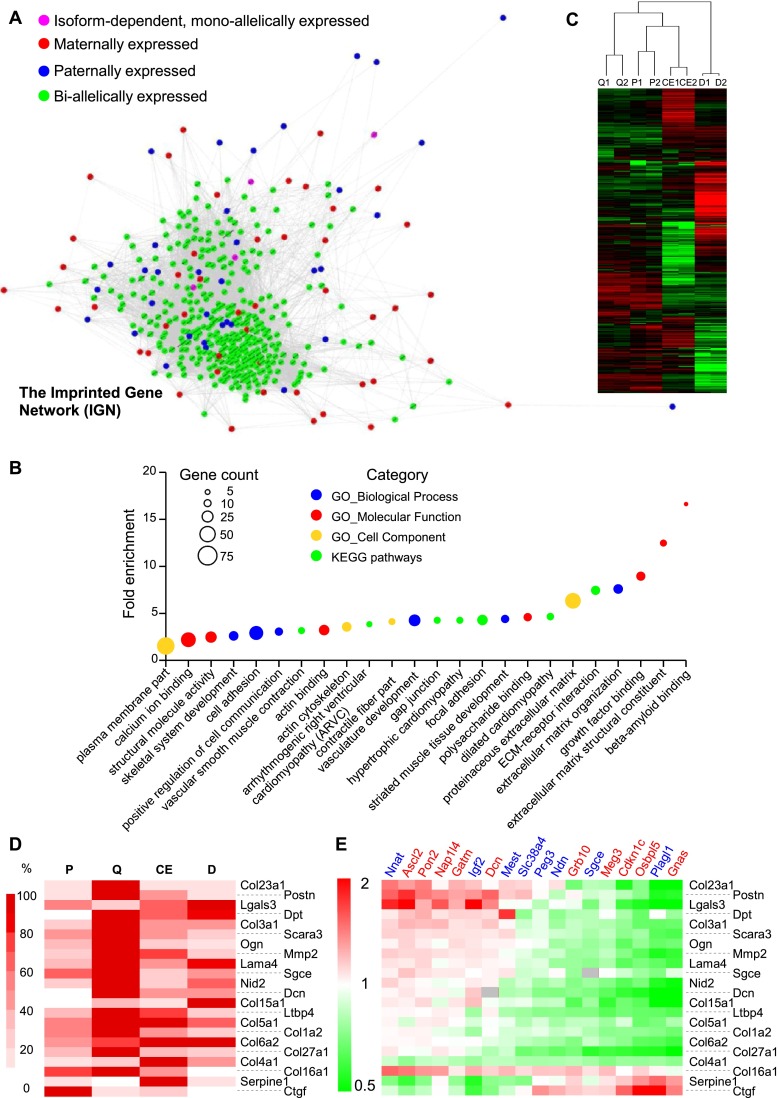
To gain insight into the mechanisms by which IGs control proliferation and differentiation, we searched COXPRESdb and Gemma for genes coexpressed with murine IGs. We ranked these genes to select those most tightly coexpressed with a maximal number of IGs. We then intersected the two ranked lists of biallelically expressed genes (bi-Gs) and identified 324 genes, which together with 85 IGs, constitute the whole murine IG network (IGN) (Fig. 6A; Supplemental Table S10; for details regarding the construction of the IGN, see Supplemental Information). To identify the biological process(es) associated with this network, we tested the overrepresentation of GO terms and KEGG pathways. The IGN was significantly enriched with genes that encode extracellular matrix (ECM) and actin fiber constituents and that are involved in cell adhesion, cell junction, and ECM-activated and growth factor–activated signaling (Fig. 6B; Supplemental Table S11). To test whether the overrepresentation of these terms is significant or due to a bias in Gemma and COXPRESdb, e.g., because of overrepresentation of some biological conditions, we constructed “networks” with 25 sets of randomly selected genes using the same procedure. For each of the 25 resulting networks, we used the DAVID tools to identify terms significantly enriched (Supplemental Table S12). This resulted in 592 terms significantly overrepresented in the 25 networks. The distribution of the number of enriched terms per network was wide (median, 12; interquartile range, 26; total range, 0–128), as was expected for random networks made of poorly connected, small sets of connected genes. Of 592 enriched terms, only three (0.5%) were found among terms associated with the genes that constitute the IGN. We found an “extracellular matrix” in a GO_BP (GO:0030198;extracellular matrix organization) and a GO_MF (GO:0005201;extracellular matrix structural constituent) and “adhesion” in a KEGG pathway (mmu04510:Focal adhesion). Other terms enriched in the IGN were absent from the 592 terms enriched in the 25 random networks. The three enriched terms were all found in the same random network (#12) out of the 25 tested. Interestingly, we retrieved the 28 genes that led to enrichment of these three terms and noticed that 14 were also IGN members. These 14 genes constituted 50%, 66%, and 61% of the genes that led to overrepresentation of the three terms (see highlighted terms in Supplemental Table S12). Our conclusion was that terms enriched in the IGN were generally not found among terms enriched in comparable random networks, unless the bait genes “captured” a portion of the IGN during the network construction process. If this portion was large enough, the corresponding network may display an enrichment of terms associated with the IGN such as ECM organization and focal adhesion.
Figure 6.
The murine imprinted gene network (IGN). (A) Biallelically expressed genes coexpressed with murine IGs were retrieved from COXPRESdb and Gemma. Coexpression links among genes present in the intersection of the two lists (for details, see Supplemental Information) were retrieved from COXPRESdb and represented using Cytoscape. Node size and edge width do not map numerical data. (B) GO terms and KEGG pathways enriched in the set of genes represented in Figure 6A. The fold enrichment is displayed for GO terms/KEGG pathways with Benjamini-Hochberg–corrected P-values < 0.05. The size of each dot is proportional to the number of genes associated with the corresponding GO term/KEGG pathway. (C) Hierarchical clustering of transcriptome data from 3T3-L1 preadipocytes. The transcriptome of two independent cultures of 3T3-L1 cells during exponential growth (P; 48 h prior confluence), quiescence (Q; 48 h post-confluence), the clonal expansion phase (CE; 10 h following addition of IDX), and in the differentiated state (D; 6 d following addition of IDX) was determined using RNA-seq. (D) Heatmap of normalized RNA-seq counts for selected ECM genes. (E) Effect of IG overexpression on ECM gene expression. Complementary DNAs encoding CAT or various IGs were transfected in exponentially growing 3T3-L1 cells. Three days post-transfection, the ECM genes were quantified using real-time PCR, and the ratio to the expression levels in the control condition (CAT) was calculated for each IG and represented as a heatmap. Dcn and Sgce are imprinted ECM genes whose expression levels are not displayed (gray boxes) in the corresponding transfected cells.
We also searched Gemma and COXPRESdb for genes coexpressed with human IGs using the same procedure as the one described for murine IGs. We identified 275 biallelically expressed genes coexpressed with the 69 human IGs present in both Gemma and COXPRESdb. As for the murine IGN, we drew the corresponding human IGN using the coexpression links from COXPRESdb (Supplemental Fig. S13A; Supplemental Table S13). Interestingly, the terms overrepresented among the genes of this network were again related to the ECM, cell adhesion, ECM-receptor interaction, focal adhesion, and cell communication (Supplemental Fig. S13B; Supplemental Table S14).
To verify that our in silico observations were relevant in cellulo, we tested whether the bi-Gs that constitute the murine IGN were indeed enriched in genes coexpressed with IGs in the 3T3-L1 adipogenic differentiation model. We performed RNA-seq on duplicate cultures of proliferating (P), quiescent (Q), clonal expansion (CE), and differentiated (D) 3T3-L1 cells (Fig. 6C). As expected, IGs and IGN members were enriched among genes that were statistically more abundantly expressed in Q compared to P and CE, whereas they were not, or even depleted, in genes that were less abundantly expressed in Q compared to P and CE (Supplemental Table S15). GO terms and KEGG pathways associated with genes up-regulated in Q compared to P and CE (Supplemental Fig. S12A; Supplemental Table S16) were very similar to those overrepresented in the IGN (Fig. 6B). In contrast, genes significantly down-regulated in Q compared to P and CE were enriched in a completely different set of GO terms and KEGG pathways (Supplemental Fig. S12B; Supplemental Table S17). These results indicated that a significant fraction of the IGN genes was induced when preadipocytes exited the cell cycle and repressed when quiescent cells resumed proliferation, supporting the existence of the IGN in cellulo.
Finally, we tested whether IGs modulated ECM gene expression. We selected 22 ECM genes that were dynamically regulated in the adipogenic differentiation model (Fig. 6D). We transfected proliferating preadipocytes with cDNAs encoding IGs as above and monitored ECM gene expression when the cells were quiescent (Supplemental Fig. S14). Interestingly, overexpression of most IGs resulted in altered expression of ECM genes (Fig. 6E). At least two groups of IGs could be distinguished according to their effect on the expression of ECM genes. IGs such as Nnat, Ascl2, Pon2, Nap1l4, and Gatm up-regulated most ECM genes, whereas other IGs, i.e., Gnas, Plagl1, Osbpl5, Cdkn1c, Meg3, Sgce, Grb10, Ndn, and Peg3, had the opposite effect. Moreover, two ECM genes, Serpine1 and Ctgf, whose endogenous pattern of expression was most dissimilar to that of IGs, displayed the exact reverse behavior (Fig. 6E).
Discussion
Parental genomic imprinting has commanded attention over the past three decades as a prototypical epigenetic gene regulation mechanism. The identification of mechanisms governing parent-of-origin-dependent, monoallelic expression of IGs has been instrumental in understanding some of the basic concepts in epigenetics (Barlow 2011). However, a holistic analysis of IG function has been lacking to date, and most often, IGs are functionally characterized without taking into account their imprinting status. The paradox in genomic imprinting so far is that a single mechanism was selected during evolution to regulate seemingly unrelated genes. The present work shows for the first time that they belong to a single gene network. The implementation of genomic imprinting during mammalian evolution may therefore have been to influence the biological process(es) controlled by this network as a whole.
Following early observations suggesting coregulation of a small number of IGs (Hayashida et al. 1997; Yan et al. 2003), we showed that a subset of 15 IGs is frequently coexpressed (Varrault et al. 2006), which was confirmed by other groups (Lui et al. 2008; Berg et al. 2011; Zacharek et al. 2011). The present study identified significantly more coexpression among IGs by using broader and more varied data sets; e.g., the Gemma and COXPRESdb databases comprise data sets from the nervous system, which was underrepresented in the TMM database originally explored. Networks identified through transcriptomic data may result from cell- or tissue-specific subnetworks and should thus be utilized only for sets of genes that are expressed in the same cell types. Despite this shortcoming, we verified that coregulation of IGs and IGN members was not an in silico artifact but was observed in cellulo and in vivo. Further experiments will reveal whether the IGN is invariant across all cell types or whether there is a core of coregulated IGs and biallelically expressed genes and a subset of IGN members that are coexpressed only in specific cell types.
The phenotypes of murine/ovine IG mutants and human patients with syndromes resulting from imprinting defects point to a major role for genomic imprinting during embryogenesis and post-natal development. The mechanism by which IGs impact organ size is, however, not fully understood. It was proposed that IGs act at multiple levels to control energy homeostasis (Charalambous et al. 2007; Radford et al. 2011; Peters 2014) because IGs play critical roles in the development and function of key metabolic organs: brain, pituitary, adrenal, pancreas, muscle, white and brown adipose tissue, and liver. In this view, each IG is proposed to have a key function in the tissue(s) most affected by the corresponding mutant. The control of organismal growth would be accomplished by a whole set of seemingly diverse processes, as suggested by the lack of common GO terms associated with IGs. Our work suggests an alternative mechanism in which IGs cooperatively control a single biological process, which impacts the function of various organs involved in the control of organismal growth. Both views are, however, easily reconcilable as different IGs cooperating in the same network may have a limiting function in different cell types. In line with this hypothesis, Hippenmeyer et al. (2013) demonstrated that uniparental disomy of paternal Chromosome 7, and in particular Igf2, caused paternal growth dominance in the liver and the lung but not in the brain and the heart. Other processes such as tumor formation (Holm et al. 2005), reprogramming of somatic cells to induced pluripotent stem cells (Stadtfeld et al. 2010), and adaptation to post-natal life (Plagge et al. 2004; Charalambous et al. 2012) were shown to be affected by deregulation of several IGs. Despite the apparent unrelatedness of these processes, they all involve the precise control of the balance between cell proliferation, quiescence, and differentiation. The demonstration that IGs are involved in the transition between proliferating and quiescent/differentiated cells provides a rationale for their observed role in these processes.
It is noteworthy that in all tests depicted in Figure 5, maternally and paternally expressed IGs did not systematically display antagonistic effects. This argues against the view that maternally expressed IGs restrict growth because they favor quiescence and differentiation, whereas paternally expressed IGs enhance growth by favoring proliferation. The link between the functional properties of IGs at the cellular and organismal levels is however complex, as exemplified by Plagl1 nullizygotes (Varrault et al. 2006). Plagl1 is paternally expressed, and in agreement with the kinship theory of imprinting (Moore and Haig 1991), Plagl1+/−pat and nullizygotes display embryonic growth restriction. However, at the cellular level, PLAGL1 displays an antiproliferative activity (Spengler et al. 1997). Furthermore, in Plagl1 nullizygotes, liver is hypoplastic (Varrault et al. 2006), whereas the retina displays amacrine cell hyperplasia (Ma et al. 2007). Other published data argue against a simplistic interpretation of our results. For instance, the Angelman and Prader-Willi syndromes result from the loss of reciprocally IGs and yet display common phenotypes such as obesity, i.e., hyperplastic and/or hypertrophic adipose tissue. Similarly, Igf2, whose role as a growth factor is supported by an abundant literature, is induced in quiescent versus proliferating cells and expressed at maximal levels when cells exit the cell cycle to differentiate (see Fig. 2D). These examples indicate that the link between the molecular, cellular, and organismal properties of IGs is complex; our results demonstrate that at the cellular level, IGs control the transition between proliferating and quiescent/differentiated states. This discovery will certainly help to link the molecular and organismal levels of analysis.
In addition to IGs, the IGN includes numerous biallelically expressed, nonimprinted genes. GO terms and KEGG pathways analysis revealed that the IGN is enriched in genes involved in cell communication, e.g., the TGF beta, WNT, IGF, and BMP pathways, which was not unexpected since several IGs are themselves involved in this biological process. In contrast, the abundance of genes involved in the ECM organization; ECM-receptor interactions; cell adhesion, including focal adhesion and gap junction; and the control of the actin cytoskeleton was unexpected. In addition, IGN members are not enriched in cell cycle genes. These data suggest that the IGN ultimately regulates proliferation and cell cycle exit but does not directly impact the cell cycle machinery. The nature of the IGN members is rather suggestive of an indirect role of this network on proliferation-quiescence-differentiation through the modulation of the ECM composition. The influence of the ECM on cellular fate is best studied in the context of the stem cell niche (Brizzi et al. 2012), where it controls several parameters known to affect stem cell behavior, including mechanical stiffness, growth factors and morphogens availability, and cell-matrix anchorage. Our observations suggest that IGs may control the behavior of stem cells through the modulation of the ECM composition. Indeed, several IGs, including Ascl2 (previously known as Mash2) (van der Flier et al. 2009), Cdkn1c (Bilodeau et al. 2009; Matsumoto et al. 2011; Zou et al. 2011; Mairet-Coello et al. 2012; Furutachi et al. 2013), Dlk1 (Ferrón et al. 2011; Mirshekar-Syahkal et al. 2013), H19 and Igf2 (Bracko et al. 2012; Venkatraman et al. 2013), Ndn (Kubota et al. 2009; Asai et al. 2012), and others (Zacharek et al. 2011), were recently suggested to control adult and embryonic stem cell fate; in particular, self-renewal, proliferation, and/or differentiation. The IG expression data in the muscle regeneration model suggested that it may also be a general property of IGs in vivo.
The demonstration that IGs are part of a single gene network that also includes biallelically expressed, nonimprinted genes is also of interest with respect to parent-of-origin effects in mammals and implementation of genomic imprinting during evolution. Mott et al. (2014) recently reported that a surprisingly large proportion (93%) of quantitative traits in mouse displayed parent-of-origin effects. They provided genetic evidence that nonimprinted genes can generate parent-of-origin effects by interacting with imprinted loci and deduced that the importance of the number of IGs is secondary to their interactions. In this context, the description of the IGN provides a molecular substratum for the genetic effects observed by Mott et al. (2014) and further highlights the importance of the IGN for many physiological traits.
Our work did not provide support to nor rule out any of the different theories of genomic imprinting, whether they put forward the conflict of parental genomes (Moore and Haig 1991), the prevention of parthenogenesis, the maternal–fetal coadaptation, and the genome defense (for a review of “nonconflict” hypotheses, see Spencer and Clark 2014). Yet, at the cellular level, we did not find evidence of antagonistic activities of paternally and maternally expressed genes, which does not support the conflict theory, although the link between the cellular and organismal levels is not straightforward, as pointed out above. The identification of the IGN suggests that genomic imprinting was not created ex nihilo, but rather targeted the regulatory genes of a preexisting machinery. In that view, genomic imprinting was not implemented to control the transition from proliferation to quiescence/differentiation but to adapt this biological process to some challenge(s) specific to therian mammals. Rather than providing a novel hypothesis about the logic behind the selection of genomic imprinting during mammalian evolution, our work provides a frame to understand how genomic imprinting was implemented and developed. In marsupials, the first clade displaying genomic imprinting in animals, the number of IGs is limited. Wolf (2013) recently suggested that the presence of gene interactions, which favor genetic coadaptation, could also favor the evolution of genomic imprinting. This hypothesis and the identification of the IGN suggest how genomic imprinting of a minimal number of genes in marsupials led to its “spreading” across a preexisting network of interacting genes in eutherian mammals. From a more physiological point of view, the increase in the number of IGs from metatherian to eutherian mammals might reflect the increased complexity of extended embryonic development in eutherians compared to metatherians.
Methods
Mining meta-analyses of microarray data and bioinformatics
We retrieved the 124 murine IGs (Supplemental Table S1) from GeneImprint (http://www.geneimprint.com), the Wamidex atlas (https://atlas.genetics.kcl.ac.uk/), and the Harwell Mousebook Imprinting Catalog (http://www.mousebook.org). We searched COXPRESdb (http://coxpresdb.hgc.jp) for murine IG data and retrieved the genes coexpressed with each of the 85 IGs present in this database. We then filtered these lists for coexpression links that involve two IGs with a mutual rank below 1200 (for a discussion of this parameter, see Supplemental Information). The resulting network was drawn using Cytoscape (Shannon et al. 2003). To generate random murine networks from COXPRESdb, we randomly selected 10,000 sets of 85 GeneIDs from the list of genes present in this database. We retrieved coexpression links as above and calculated the average degree and average mutual rank. We performed the same analysis with sets of 85 genes that belonged to Gene Ontology Biological Processes (GO_BPs). We selected 50 GO_BPs based only on the number of genes with data in COXPRESdb they comprised, i.e., at least 85. In case the GO_BPs included more than 85 genes with data in COXPRESdb, we used the 85 first GeneIDs in the list of genes found in the corresponding GO_BP. To identify genes coexpressed with murine IGs in Gemma (http://www.chibi.ubc.ca/Gemma/home.html), we retrieved the genes coexpressed with each of the 101 IGs present in this database. We then filtered these lists for coexpression links that involve two IGs in a minimum of two data sets. The resulting network was drawn using Cytoscape. The same approach was performed with the 86 human IGs present in Gemma and the 60 rat IGs and murine IG orthologs present in COXPRESdb. Human IGs were retrieved from GeneImprint and from the Catalogue of Imprinted Effects (http://igc.otago.ac.nz/home.html) (Morison et al. 2005). For the rat network, orthologs of murine IGs were retrieved from Homologene and manually curated. The construction of the imprinted gene network is described in detail in the Supplemental Information. Network analysis was performed using the StatsBase, Distributions, and Graphs packages in Julia (http://julialang.org/), as well as custom scripts.
Gene functional classification
Functional analyzes of gene lists were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID 6.7; 2013-01-19) tools with the threshold count and EASE factor set at two and 0.1, respectively (Huang et al. 2009).
Cell culture
All cells were grown in DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin in a humidified atmosphere containing 5% CO2 at 37°C. Mouse embryonic fibroblasts from C57BL/6J mouse strain were prepared using standard protocols (Xu 2005). For serum deprivation, the cells were washed twice with 1× PBS, and fresh medium containing 0.1% FBS was added for the indicated period of time. 3T3-L1 preadipocytes were from ATCC (CL-173). For maintenance, the medium was changed every 2 d, and the cells were always kept at <15% of the saturating cell density. Cells were split a maximum of four rounds. For differentiation, cells were seeded at low density and grown to confluence. After 2 d at confluence, adipogenic differentiation was initiated by addition of insulin (5 mg/mL, Sigma), dexamethasone (1 μM, Sigma), and isobutylmethylxanthine (IBMX; 250 μM, Sigma). Medium was then renewed every 2 d with insulin only. Cell counts were determined using a CASY TT apparatus (Innovatis, Roche).
Muscle regeneration
All animal procedures were conducted in accordance with the protocol approved by the Institutional Animal Care and Use Committee. Notexin solution (10 μL at 50 μg/mL in 0.9% NaCl, Sigma) was injected into the left tibialis anterior of male C57BL/6J mice (79 ± 5 d); saline-injected muscles were used to monitor the absence of significant gene regulation in the absence of notexin, not to normalize data. Two mice were killed at each time point, and the injected tibialis anterior was removed and homogenized in RNA-Now (Ozyme). Levels of expression of proliferation, inflammation, and myogenesis markers and IGs were measured by real-time RT-PCR using Mrpl32 as a reference gene.
Parent-of-origin-dependent gene expression
Mouse embryonic fibroblasts from E13.5 embryos were prepared as above from two reciprocal crosses, BJ1 (♀C57BL/6J × ♂JF1/Ms) and JB6 (♀JF1/Ms × ♂C57BL/6J), and grown in vitro. Genomic DNA and RNA were prepared from MEFs at different cell densities. To monitor parent-of-origin-dependent expression, genomic DNA and cDNA were PCR amplified using primers surrounding the polymorphic region: nt 1330 on NM_010052 for Dlk1, nt 8540–8779 on NM_010515 for Igf2r, nt 1597–1884 on NR_027652 for Meg3, and nt 691–1511 on NR_001592 for H19. Depending on the nature of the polymorphism, three techniques were used: (1) visualization of the amplicon size by agarose gel electrophoresis as for Igf2r, (2) analysis of RFLP as for Meg3 using Bsh1236I and H19 using BclI, and (3), direct sequencing of the amplicon as for Dlk1. Primers were Dlk1HybF (CGTCTTTCTCAACAAGTGCG), Dlk1HybR (AGATCTCCTCATCACCAGCC), H19HybF (GGATCCAGCAAGAACAGAAGC), H19HybR (TCTGTCCTCTCCATCACACC), Meg3HybF (TTGCACATTTCCTGTGGGAC), Meg3HybR (AAGCACCATGAGCCACTAGG), Igf2rHybF (TTCGACCTATAAGAAGCCTT), and Igf2rHybR (GGGTACTTTGCTTTTGGGTA).
Methylation pattern of differentially methylated regions at IG loci
Genomic DNA was prepared with the PureLink genomic DNA kit (Invitrogen). One microgram of genomic DNA in 32.5 μL of 1× PBS was denatured for 15 min at 50°C in the presence of NaOH (0.3 M final). This solution was supplemented with hydroquinone (75 mM final) and sodium pyrosulfate (4 M final), both prepared extemporaneously. DNA was incubated 1 min at 95°C and 4 h at 55°C. DNA was desalted and purified using Zymo-Spin IC columns (Zymo Research). Ten nanograms of bisulfite-treated DNA were PCR amplified using OneTaq DNA Polymerase (New England Biolabs). Amplicons were purified, cloned using Zero Blunt TOPO PCR cloning kit (Invitrogen), and sequenced. The identification of methylated cytosine was performed using CpGviewer (Carr et al. 2007). Sequences of primers to amplify bisulfite-treated genomic DNA (Tomizawa et al. 2011) are given in Supplemental Table S18.
Transfection
Exponentially growing 3T3-L1 cells were maintained at a maximum density of 7500/cm2 (∼10% of saturation density) and transfected by electroporation (Neon, Life Technologies) according to the supplier’s recommendations. Cells were detached with trypsin/EDTA at 37°C. After centrifugation, the cells were resuspended in buffer R (Life Technologies) to 10 M cells/mL. One hundred microliters of the suspension was mixed with plasmids/siRNA, electroporated (two pulses, 20 msec, 1300 V), resuspended in the appropriate volume of DMEM+10% FBS without antibiotics, and seeded at the desired density. The medium was renewed after 24 h with standard medium. For cDNA transfection, 1 μg of pRK5-eGFP was mixed with 4 μg pRK5-IG per million cells. For siRNA transfection, 2 μL of siRNA at 100 μM was used per million cells. The final concentration of siRNA in the medium varies, depending on experience, between 20 and 50 nM.
cDNAs
cDNAs encoding eGFP, CAT, and IGs were subcloned into pRK5 and derivatives (Supplemental Table S19) and verified by sequencing. Plasmid pRK5-CD2 was described previously (Bilanges et al. 2001).
Small interfering RNAs
Two scramble siRNAs, which do not affect the proliferation and differentiation of 3T3-L1 cells (data not shown), were used as negative controls. The efficiency of each siRNA was evaluated by measuring the residual expression of the target transcript by RT-qPCR (Supplemental Fig. S9). Only siRNAs resulting in at least 70% knockdown compared with scramble siRNAs were selected (Supplemental Table S20). To ensure the specificity of the loss-of-function experiments, two different siRNAs and one esiRNA (see infra) for most IG were validated and used separately. For Ndn and Sgce, Peg3 and Peg10, and Mest, two, three, and four siRNAs, respectively, were used because no esiRNA was available or could be validated. The siRNAs (GeneCust, Luxembourg) were methylated on position 2 of the ribose of the first and second nucleotides of the sense strand and on the first one of the antisense strand in order to avoid off-target effects (Jackson and Linsley 2010; AL Jackson, pers. comm.). In addition, we used one esiRNA (endoribonuclease-prepared siRNA; MISSION esiRNA, Sigma), which consists of a mixture of different siRNA molecules generated from a transcribed sequence. The total siRNA concentration is effective on the target gene, but each unique sequence is present at a low concentration, which minimizes off-target effects. The phenotypes we obtained were similar for the selected siRNAs, and Figure 5 displays the average data obtained with the two to four different siRNAs/esiRNAs per IG.
Adipogenic differentiation
Oil Red O (ORO; Sigma #00625) solution was prepared in isopropanol (1.8 mg/mL) and filtered. Differentiated cells were fixed in 4% formaldehyde for 30 min and then washed in 1× PBS. Fixed cells were stained in 60% ORO stock solution for 30 min at 37°C and then washed in isopropanol. For ORO staining quantification, ORO was dissolved in DMSO (Sigma), and the optical density was measured at 510 nm. Determining adipogenic differentiation by flow cytometry was performed as previously described (Lee et al. 2004b) with some changes. Cells were rinsed twice with 1× PBS and detached with prewarmed 0.25% trypsin-EDTA. Trypsin was inactivated by the addition of three volumes of cold PBS-10% FCS and kept on ice prior to flow cytometric analysis. Side scatters (SSC) and forward scatters (FSC) were set so that 99.5% of control cells (quiescent cells before induction) were negative. When GFP+ populations were studied, 5,000 GFP+ cells were analyzed.
BrdU incorporation
Cells were pulsed for 30 min with 20 μM BrdU, washed with 1× PBS, trypsinized, centrifuged, and fixed in ethanol 70%. Samples were kept at −20°C for at least 48 h prior to staining. Fixed cells were centrifuged to remove ethanol and rehydrated in PBS-0.5% BSA. After centrifugation, the cells were resuspended in 200 μL Tris-NaCl (pH 7.8) per million cells and treated with 1 mL 2.5 M HCl per million cells for 20 min. HCl was diluted with the same volume of PBS-0.5% BSA. After centrifugation, residual HCl was neutralized with 0.1 M borate buffer (pH 8.5). The cells were washed twice with PBS-0.5% BSA. Cell pellets were resuspended in 10 μL of FITC-labeled anti-BrdU (Roche) diluted 1/20 and incubated for 30 min at 4°C. The cells were washed twice in PBS-0.5% BSA and resuspended in the same solution. The cytometric analysis was performed with a FACS Calibur II (BD Biosciences).
Real-time RT-PCR
Total RNA was prepared using RNA Now (Ozyme), treated with DNase (DNA-free, Ambion), and reverse-transcribed with MoMuLV-RT (Life Technology) according to manufacturer’s protocols. To quantify the expression of imprinted and ECM genes as well as proliferation, adipogenesis, myogenesis, and inflammation markers, we designed and validated a collection of primer pairs (Supplemental Table S21) to be used in real-time, quantitative RT-PCR, which was performed with Roche’s LightCycler 480 and LC480 SYBR green I master reagent. The level of expression of each gene “X” was normalized to the geometric mean of the expression levels of the selected reference genes, R1 to R3, in the same PCR plate according to the formula:
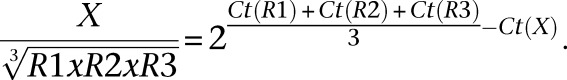 |
Reference genes were selected using the GeNorm procedure (Vandesompele et al. 2002). For the muscle regeneration experiments, only Mrpl32 was found to be stable across all samples and used for normalization.
RNA-seq
Libraries were generated using Illumina’s TruSeq RNA sample prep kit v2 (RS-122-2001) from two independent experiments. Briefly, polyadenylated RNAs were isolated from 600 ng of total RNA using oligo(dT) magnetic beads. Poly(A)+ RNA were fragmented, primed using random hexamer, and reverse transcribed. The second-strand synthesis was performed, and double-stranded cDNAs were blunt ended. 3′ ends were adenylated and ligated to Illumina’s indexed adapters. Each library was PCR amplified (15 cycles) using Illumina’s PCR primers. Libraries were validated using a DNA 1000 chip (Bioanalyzer; Agilent) and quantified by qPCR using the Library Quantification Kit-Illumina/Universal from Kapa Biosystems (KK4824). Prior to clustering, libraries were denatured using NaOH and diluted to 8 pM. Clustering and paired ends (2 × 100 nt) sequencing on a HiSeq 2000 were performed on a single lane per library following the manufacturer’s instructions. In this experiment, only reads #1 were used and mapped to the full mm9 mouse reference genome using the eland_rna module of CASAVA 1.8.2 (Illumina); 118.3–160.7 M reads per library mapped uniquely to the mouse transcriptome. Counting was performed with CASAVA’s variant detection and counting module. UCSC annotation file (refFlat.txt.gz; 2012-01-15) was used to retrieve annotations on genes and exons. GeneIDs are from the NCBI (gene2refseq.gz; 2012-01-19). From the 19,489 genes with at least one count in one sample, 3305 genes for which the sum of raw counts in the eight samples was less than 20 were filtered out. Counts were normalized using the RLE method as implemented in edgeR (Robinson et al. 2010), which was used to identify differentially expressed genes with a generalized linear model likelihood ratio test using a paired design and a FDR set at 1%. Centroid linkage clustering of RNA-seq data, samples and genes was performed with Cluster 3.0 using uncentered Pearson correlation as distance (de Hoon et al. 2004).
Statistics
All data are presented as mean ± SEM unless otherwise specified. Cell culture experiments were repeated at least three independent times. Statistical analysis of cell culture experiments was performed using Prism (GraphPad Software) with a Kruskal-Wallis test followed by two-tailed Wilcoxon signed rank tests with Benjamini-Hochberg correction for multiple testing.
Data access
The RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) (Edgar et al. 2002) under accession number GSE50612.
Supplementary Material
Acknowledgments
We thank Paul Pavlidis, Anton Zoubarev, and Patrick Tan for help with the analysis of the Gemma data, Annie Delalbre and Gilles Carnac for help with notexin injection experiments, Robert Feil and Satya K. Kota for providing JB6 and BJ1 MEFs and primer sequences for the McrBC analysis, Philippe Arnaud and Atsuo Ogura for providing information about SNPs in murine IGs, Julien Cau and Amélie Sarrazin (MRI imaging and cytometry facility) for cell cytometry, Christelle Reynès and Robert Sabatier for advice with statistical analysis of the data, Vincent Demolombe for bioinformatic support, and William Ritchie, Gavin Kelsey, and Marisa S. Bartolomei for critical reading of the manuscript. This research was supported by grants from the CNRS, INSERM, Montpellier University, and the National Research Agency (ANR-07BLAN052). H.A.A., B.E., and C.G. were supported by fellowships from the Ministry for Higher Education and Research. B.E. and C.G. were supported by fellowships from La Ligue contre le Cancer and L’Association pour la Recherche contre le Cancer, respectively. The MGX-Montpellier GenomiX facility, a member of the “France Génomique” network, is supported by the Languedoc-Roussillon district and the National Research Agency (ANR-10-INBS-09-06).
Author contributions: H.A.A., T.B., A.V., and L.J. conceived and designed the experiments. H.A.A., B.E., A.L.D., C.G., H.P., T.B., A.V., and L.J. performed the experiments. H.A.A., B.E., A.L.D., C.G., E.D., T.B., A.V., and L.J. analyzed the data. C.D. contributed analysis tools. H.A.A., B.E., T.B., A.V., and L.J. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.175919.114. Freely available online through the Genome Research Open Access option.
References
- Asai T, Liu Y, Di Giandomenico S, Bae N, Ndiaye-Lobry D, Deblasio A, Menendez S, Antipin Y, Reva B, Wevrick R, et al. . 2012. Necdin, a p53 target gene, regulates the quiescence and response to genotoxic stress of hematopoietic stem/progenitor cells. Blood 120: 1601–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow DP. 2011. Genomic imprinting: a mammalian epigenetic discovery model. Annu Rev Genet 45: 379–403. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Ferguson-Smith AC. 2011. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol 3: a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Lin KK, Sonnet C, Boles NC, Weksberg DC, Nguyen H, Holt LJ, Rickwood D, Daly RJ, Goodell MA. 2011. Imprinted genes that regulate early mammalian growth are coexpressed in somatic stem cells. PLoS ONE 6: e26410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilanges B, Varrault A, Mazumdar A, Pantaloni C, Hoffmann A, Bockaert J, Spengler D, Journot L. 2001. Alternative splicing of the imprinted candidate tumor suppressor gene ZAC regulates its antiproliferative and DNA binding activities. Oncogene 20: 1246–1253. [DOI] [PubMed] [Google Scholar]
- Bilodeau S, Roussel-Gervais A, Drouin J. 2009. Distinct developmental roles of cell cycle inhibitors p57Kip2 and p27Kip1 distinguish pituitary progenitor cell cycle exit from cell cycle reentry of differentiated cells. Mol Cell Biol 29: 1895–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracko O, Singer T, Aigner S, Knobloch M, Winner B, Ray J, Clemenson GD Jr, Suh H, Couillard-Despres S, Aigner L, et al. . 2012. Gene expression profiling of neural stem cells and their neuronal progeny reveals IGF2 as a regulator of adult hippocampal neurogenesis. J Neurosci 32: 3376–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzi MF, Tarone G, Defilippi P. 2012. Extracellular matrix, integrins, and growth factors as tailors of the stem cell niche. Curr Opin Cell Biol 24: 645–651. [DOI] [PubMed] [Google Scholar]
- Carr IM, Valleley EMA, Cordery SF, Markham AF, Bonthron DT. 2007. Sequence analysis and editing for bisulphite genomic sequencing projects. Nucleic Acids Res 35: e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattanach BM, Kirk M. 1985. Differential activity of maternally and paternally derived chromosome regions in mice. Nature 315: 496–498. [DOI] [PubMed] [Google Scholar]
- Cattanach BM, Beechey CV, Peters J. 2006. Interactions between imprinting effects: summary and review. Cytogenet Genome Res 113: 17–23. [DOI] [PubMed] [Google Scholar]
- Charalambous M, da Rocha ST, Ferguson-Smith AC. 2007. Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr Opin Endocrinol Diabetes Obes 14: 3–12. [DOI] [PubMed] [Google Scholar]
- Charalambous M, Ferron SR, da Rocha ST, Murray AJ, Rowland T, Ito M, Schuster-Gossler K, Hernandez A, Ferguson-Smith AC. 2012. Imprinted gene dosage is critical for the transition to independent life. Cell Metab 15: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. 2004. Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith AC. 2011. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet 12: 565–575. [DOI] [PubMed] [Google Scholar]
- Ferrón SR, Charalambous M, Radford E, McEwen K, Wildner H, Hind E, Morante-Redolat JM, Laborda J, Guillemot F, Bauer SR, et al. . 2011. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature 475: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S, Matsumoto A, Nakayama KI, Gotoh Y. 2013. p57 controls adult neural stem cell quiescence and modulates the pace of lifelong neurogenesis. EMBO J 32: 970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Kehinde O. 1974. Sublines of 3T3 cells that accumulate lipids. Cell 1: 113–116. [Google Scholar]
- Hayashida T, Eversole-Cire P, Jones PA, Sasaki H. 1997. Imprinted genes are up-regulated by growth arrest in embryonic fibroblasts. J Biochem 122: 901–903. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Johnson RL, Luo L. 2013. Mosaic analysis with double markers reveals cell-type-specific paternal growth dominance. Cell Rep 3: 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm TM, Jackson-Grusby L, Brambrink T, Yamada Y, Rideout WM III, Jaenisch R. 2005. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell 8: 275–285. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Linsley PS. 2010. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov 9: 57–67. [DOI] [PubMed] [Google Scholar]
- Kelsey G, Feil R. 2013. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos Trans R Soc Lond B Biol Sci 368: 20110336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Osawa M, Jakt LM, Yoshikawa K, Nishikawa SI. 2009. Necdin restricts proliferation of hematopoietic stem cells during hematopoietic regeneration. Blood 114: 4383–4392. [DOI] [PubMed] [Google Scholar]
- Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. 2004a. Coexpression analysis of human genes across many microarray data sets. Genome Res 14: 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Chen SY, Wiesner RJ, Huang YF. 2004b. Simple flow cytometric method used to assess lipid accumulation in fat cells. J Lipid Res 45: 1162–1167. [DOI] [PubMed] [Google Scholar]
- Lui JC, Finkielstain GP, Barnes KM, Baron J. 2008. An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am J Physiol Regul Integr Comp Physiol 295: R189–R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Cantrup R, Varrault A, Colak D, Klenin N, Götz M, McFarlane S, Journot L, Schuurmans C. 2007. Zac1 functions through TGFβII to negatively regulate cell number in the developing retina. Neural Dev 2: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairet-Coello G, Tury A, Van Buskirk E, Robinson K, Genestine M, DiCicco-Bloom E. 2012. p57KIP2 regulates radial glia and intermediate precursor cell cycle dynamics and lower layer neurogenesis in developing cerebral cortex. Development 139: 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Takeishi S, Kanie T, Susaki E, Onoyama I, Tateishi Y, Nakayama K, Nakayama KI. 2011. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell 9: 262–271. [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D. 1984. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37: 179–183. [DOI] [PubMed] [Google Scholar]
- Mirshekar-Syahkal B, Haak E, Kimber GM, van Leusden K, Harvey K, O’Rourke J, Laborda J, Bauer SR, de Bruijn MFTR, Ferguson-Smith AC, et al. . 2013. Dlk1 is a negative regulator of emerging hematopoietic stem and progenitor cells. Haematologica 98: 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Haig D. 1991. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet 7: 45–49. [DOI] [PubMed] [Google Scholar]
- Morison IM, Ramsay JP, Spencer HG. 2005. A census of mammalian imprinting. Trends Genet 21: 457–465. [DOI] [PubMed] [Google Scholar]
- Mott R, Yuan W, Kaisaki P, Gan X, Cleak J, Edwards A, Baud A, Flint J. 2014. The architecture of parent-of-origin effects in mice. Cell 156: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K. 2011. COXPRESdb: a database to compare gene coexpression in seven model animals. Nucleic Acids Res 39: D1016–D1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. 2014. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet 15: 517–530. [DOI] [PubMed] [Google Scholar]
- Plagge A, Gordon E, Dean W, Boiani R, Cinti S, Peters J, Kelsey G. 2004. The imprinted signaling protein XLαs is required for postnatal adaptation to feeding. Nat Genet 36: 818–826. [DOI] [PubMed] [Google Scholar]
- Radford EJ, Ferrón SR, Ferguson-Smith AC. 2011. Genomic imprinting as an adaptative model of developmental plasticity. FEBS Lett 585: 2059–2066. [DOI] [PubMed] [Google Scholar]
- Renfree MB, Hore TA, Shaw G, Graves JAM, Pask AJ. 2009. Evolution of genomic imprinting: insights from marsupials and monotremes. Annu Rev Genomics Hum Genet 10: 241–262. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer HG, Clark AG. 2014. Non-conflict theories for the evolution of genomic imprinting. Heredity (Edinb) 113: 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler D, Villalba M, Hoffmann A, Pantaloni C, Houssami S, Bockaert J, Journot L. 1997. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J 16: 2814–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. 2010. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Norris ML. 1984. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308: 548–550. [DOI] [PubMed] [Google Scholar]
- Tomizawa S, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H. 2011. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development 138: 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, et al. . 2009. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136: 903–912. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, et al. . 2006. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell 11: 711–722. [DOI] [PubMed] [Google Scholar]
- Venkatraman A, He XC, Thorvaldsen JL, Sugimura R, Perry JM, Tao F, Zhao M, Christenson MK, Sanchez R, Yu JY, et al. . 2013. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature 500: 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB. 2013. Evolution of genomic imprinting as a coordinator of coadapted gene expression. Proc Natl Acad Sci 110: 5085–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. 2005. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol 70: 28.1.1–28.1.8. [DOI] [PubMed] [Google Scholar]
- Yan Z, Choi S, Liu X, Zhang M, Schageman JJ, Lee SY, Hart R, Lin L, Thurmond FA, Williams RS. 2003. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J Biol Chem 278: 8826–8836. [DOI] [PubMed] [Google Scholar]
- Zacharek SJ, Fillmore CM, Lau AN, Gludish DW, Chou A, Ho JWK, Zamponi R, Gazit R, Bock C, Jäger N, et al. . 2011. Lung stem cell self-renewal relies on BMI1-dependent control of expression at imprinted loci. Cell Stem Cell 9: 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P, Yoshihara H, Hosokawa K, Tai I, Shinmyozu K, Tsukahara F, Maru Y, Nakayama K, Nakayama KI, Suda T. 2011. p57Kip2 and p27Kip1 cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell 9: 247–261. [DOI] [PubMed] [Google Scholar]
- Zoubarev A, Hamer KM, Keshav KD, McCarthy EL, Santos JRC, Van Rossum T, McDonald C, Hall A, Wan X, Lim R, et al. . 2012. Gemma: a resource for the reuse, sharing and meta-analysis of expression profiling data. Bioinformatics 28: 2272–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.