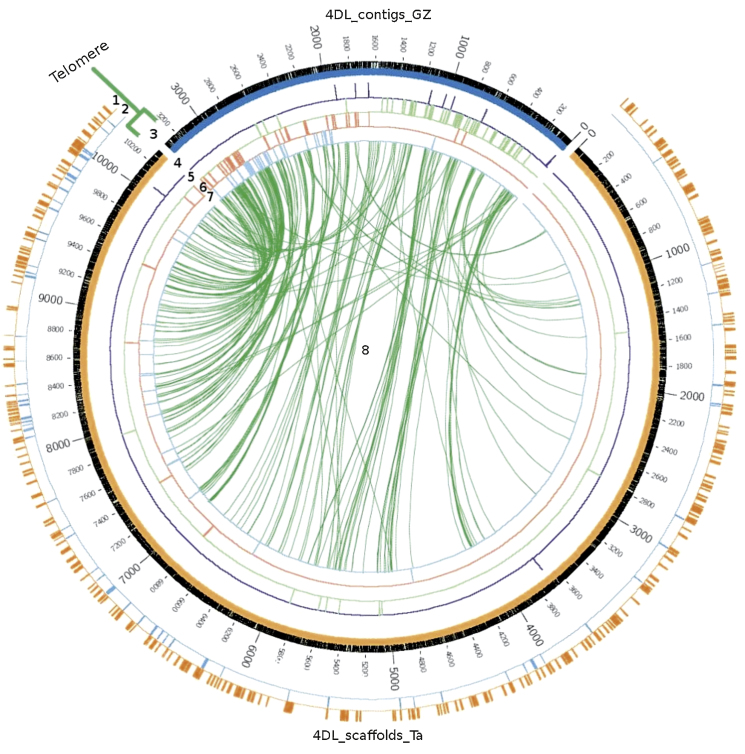
Fig. 5.
4DL GenomeZipper vs. Ae. tauschii genetic map and 4D deletion map. Comparison of 4DL scaffolds anchored and ordered according to the genetic map from Jia et al. [16] and the syntenic ordering of contigs obtained by the GenomeZipper approach (this work). Ring 1, annotated repetitive regions from scaffolds. Ring 2, annotated genes from scaffolds. Ring 3, 4DL scaffolds (orange) and GenomeZipper contigs (blue) ordered in scale where 0 represents the centromeres. Ring 4, (blue) ESTs mapped on C-4DL9-0.31 deletion bin. Ring 5, (green) ESTs mapped on 4DL9-0.31–0.56. Ring 6, (red) ESTs in 4DL13-0.56–0.71. Ring 7 (blue) ESTs in 4DL12-0.71–1.00. Ring 8, (green lines) matches between contigs ordered by GenomeZipper and scaffolds anchored to the Ae. tauschii genetic map. Deletion bin-mapping data of ESTs was obtained from Miftahudin et al. [26].