Abstract
The antimetabolite 5′-Fluorouracil (5FU) is an analog of uracil commonly employed as a chemotherapeutic agent in the treatment of a range of cancers including colorectal tumors. To assess the cellular effects of 5FU, we performed a genome-wide screening of the haploid deletion library of the eukaryotic model Schizosaccharomyces pombe. Our analysis validated previously characterized drug targets including RNA metabolism, but it also revealed unexpected mechanisms of action associated with chromosome segregation and organization (post-translational histone modification, histone exchange, heterochromatin). Further analysis showed that 5FU affects the heterochromatin structure (decreased levels of histone H3 lysine 9 methylation) and silencing (down-regulation of heterochromatic dg/dh transcripts). To our knowledge, this is the first time that defects in heterochromatin have been correlated with increased cytotoxicity to an anticancer drug. Moreover, the segregation of chromosomes, a process that requires an intact heterochromatin at centromeres, was impaired after drug exposure. These defects could be related to the induction of genes involved in chromatid cohesion and kinetochore assembly. Interestingly, we also observed that thiabendazole, a microtubule-destabilizing agent, synergistically enhanced the cytotoxic effects of 5FU. These findings point to new targets and drug combinations that could potentiate the effectiveness of 5FU-based treatments.
Keywords: Anticancer drug, chromosome segregation, chromosome organization, centromere, histone modification, heterochromatin, Schizosaccharomyces pombe, 5′-Fluorouracil
Abbreviations
- 5FU
5′-Fluorouracil, 5FU
- H3K9me
H3 lysine 9 methylation
- FdUMP
fluorodeoxyuridine monophosphate
- G1 phase
gap 1 phase of cell cycle
- S phase
synthesis phase of cell cycle
- FdUTP
fluorodeoxyuridine triphosphate
- FUTP
fluorouridine triphosphate
- GO
Gene Ontology
- HAT
histone acetyltransferase
- HMT
histone methyltransferase
- HDAC
histone deacetylase
- HULC
histone H2B ubiquitin ligase complex
- CLRC
Clr4 methyltransferase complex
- RITS
RNA-induced transcriptional silencing
- dsRNA
double-stranded RNA
- siRNA
small interfering RNA
- RNAi
interference RNA
- RDRC
RNA-directed RNA polymerase complex
- HP1
heterochromatin protein 1
- ChIP
chromatin immunoprecipitation
- imr
innermost repeats
- cnt
central core
- CENP-A
centromere-associated protein A
- MNAse
micrococcal nuclease
- TBZ
thiabendazole
Introduction
Understanding the mechanism of action of anticancer drugs is necessary to rationally design optimal strategies to improve the effectiveness of treatments as well as to propose new synergistic combinations of chemotherapeutic agents. 5′-Fluorouracil (5FU) is one of the most commonly used drugs in the treatment of several types of cancers including breast and head and neck cancers, although its effectiveness is higher in advanced colorectal tumors.1 The cytotoxic effects of 5FU have been attributed to its active metabolites: the fluorodeoxyuridine monophosphate (FdUMP) inhibits the thymidylate synthase blocking DNA synthesis and causing G1/S cell cycle arrest, and the fluorodeoxyuridine triphosphate (FdUTP) and the fluorouridine triphosphate (FUTP) can be misincorporated into DNA and RNA, respectively, interfering with normal nucleic acid metabolism.2–5 However, the molecular mechanism underlying the cytotoxic effects of 5FU has not been completely elucidated.
Recent work from our laboratory has explored the RNA-based mode of 5FU action using the eukaryotic model Schizosaccharomyces pombe.6 In this current study, we sought to identify additional pathways that could participate in 5FU cytotoxicity. For this purpose, we used a genome-wide approach that screens the fission yeast haploid deletion library searching for genes whose deletion confers sensitivity to the drug. Unexpectedly, we observed that mutants defective in chromosome segregation or organization were hypersensitive to 5FU. Moreover, we found that 5FU affects the transcriptional state and the epigenetic histone modifications of centromeric heterochromatin, probably contributing to the defects detected in chromosome segregation. Most anticancer drug studies have focused on the identification of protein targets that modulate the response to chemotherapy; however, we provide evidence that the drug effects on the chromatin state could be itself a source of cellular damage.
Results and Discussion
Genome-wide screening for genes associated with 5FU sensitivity
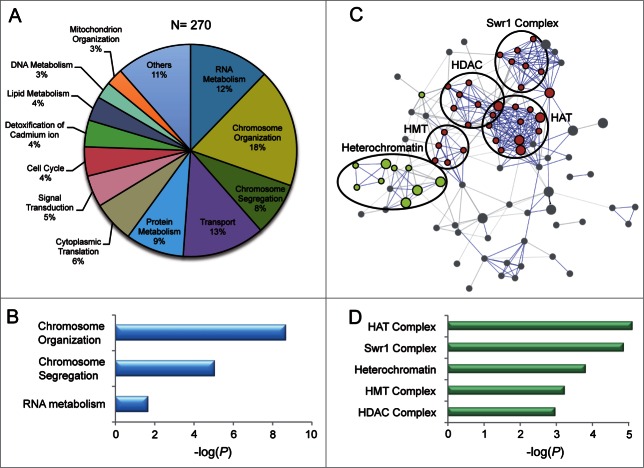
To identify novel non-essential genes whose disruption confers sensitivity to 5FU, we screened a S. pombe collection of 3,310 haploid deletion strains (Bioneer). The growth analysis revealed that 270 deletion mutants displayed severe sensitivity to 5FU in comparison with the wild-type controls (Table S1). These genes were classified according to the Gene Ontology (GO) database (http://www.geneontology.org) into several biological processes (Fig. 1A). However, statistical analysis indicated that only 3 functional categories (chromosome organization, chromosome segregation and RNA metabolism) were significantly enriched (P < 0.05) in the set of genes (Fig. 1B). The 5FU sensitivity of the strains deleted for genes involved in these categories seems to be a drug-specific effect because these mutants are statistically different from those sensitive to other drugs such as bortezomib,7 micafungin8 and terbinafine9 (Fig. S1). However, there is a significant overlap between mutants of chromosome organization and RNA metabolism genes that are sensitive to both 5FU and valproic acid (an anticancer agent that targets histone deacetylases)10 suggesting a similar mode of action (Fig. S1). The protein interaction network of over-represented groups (Fig. 1B) generated using the STRING database (http://string905.embl.de), revealed a marked enrichment in GO cellular components associated with histone modification or exchange and heterochromatin (Fig. 1C and D). Further classification of these genes into subcategories included histone acetyltransferase (HAT) complex, Swr1 complex, heterochromatin, histone methyltransferase (HMT) complex and histone deacetylase (HDAC) complex (Table 1).
Figure 1.
Classification of genes whose deletion confers sensitivity to 5FU. (A) Distribution of GO biological processes terms for the 270 genes associated with increased sensitivity to 5FU in fission yeast. (B) Negative log10 (P-values) of the GO biological processes terms calculated using the two-sided Fisher´s exact test. Only significantly over-represented groups are shown. (C) Interaction networks of the proteins grouping in chromosome organization, chromosome segregation and RNA metabolism obtained from STRING database (v9.05) using default settings (confidence > 0.4). The proteins are represented by nodes, which are colored according to the GO cellular component classification (histone modification/exchange in red and heterochromatin in green). Disconnected nodes are not shown. Lines represent the predicted functional associations between the proteins. (D) Negative log10 (P-values) evaluating the significance of the main GO cellular component terms identified in (C) in the set of 270 genes.
Table 1.
Enriched GO cellular component categories of deletion strains sensitive to 5FU. Genes were classified according to GO cellular component categories using STRING database. Note that alp13 and png2 are included in both HAT and HDAC
| Gene Name | Annotationa | Complex | Human orthologb |
|---|---|---|---|
| HAT Complex | |||
| ada2 | Transcriptional adapter 2 | SAGA | TADA2A* |
| alp13 | Chromatin modification-related protein eaf3 | nuA4 | MORF4, MORF4L1, MORF4L2 |
| bdc1 | Bromodomain-containing protein 1 | nuA4 | BRD8 |
| eaf7 | Probable chromatin modification-related protein eaf7 | nuA4 | MRGBP |
| gcn5 | Histone acetyltransferase gcn5 | SAGA | KAT2A, KAT2B |
| ngg1 | Ada histone acetyltransferase complex component | SAGA | TADA3 |
| png2 | Chromatin modification-related protein png2 | nuA4 | ING2, ING3, ING4, ING5 |
| sgf29 | SAGA-associated factor 29 homolog | SAGA | SGF29 |
| spt8 | Transcription factor spt8 | SAGA | |
| spt20 | SAGA complex subunit spt20 | SAGA | SUPT20H, SUPT20HL1, SUPT20HL2 |
| tra1 | Transcription-associated protein 1 | SAGA | TRRAP8* |
| HDAC Complex | |||
| cph2 | Clr6 histone deacetylase associated PHD protein-2 Cph2 | Rpd3 | PHF12* |
| phd1 | Histone deacetylase phd1 | Rpd3 | HDAC1, HDAC2 |
| prw1 | RbAp48-related WD40 repeat-containing protein prw1 | Rpd3 | RBBP4, RBBP7 |
| pst2 | Paired amphipathic helix protein pst2 | Rpd3 | SIN3A, SIN3B |
| set3 | SET domain-containing protein 3 | Rpd3 | KMT2E |
| rxt3 | Transcriptional regulatory protein rxt3 | Rpd3 | CRISPLD2 |
| HMT Complex | |||
| ash2 | Set1 complex component ash2 | Set1C/COMPASS | ASH2L* |
| set1 | Histone-lysine N-methyltransferase, H3 lysine-4 specific | Set1C/COMPASS | SETD1A |
| spp1 | Set1 complex component spp1 | Set1C/COMPASS | CXXC1 |
| swd1 | Set1 complex component swd1 | Set1C/COMPASS | RBBP5* |
| swd3 | Set1 complex component swd3 | Set1C/COMPASS | WDR5 |
| Swr1 Complex | |||
| arp6 | Actin-like protein arp6 | Swr1 | ACTR6* |
| bdf1 | SWR1 complex bromodomain subunit bdf1 | Swr1 | BRD2, BRD3, BRD4, BRDT |
| pht1 | Histone H2A.Z | Swr1 | H2AFV, H2AFZ |
| vps71 | SWR1 complex subunit vps71 | Swr1 | ZNHIT1* |
| swc2 | SWR1 complex protein 2 | Swr1 | VPS72* |
| swc3 | SWR1 complex subunit swc3 | Swr1 | |
| swc5 | SWR1 complex protein 5 | Swr1 | CFDP1 |
| swr1 | Helicase swr1 | Swr1 | SRCAP* |
| Heterochromatin | |||
| brl1 | E3 ubiquitin-protein ligase brl1 | HULC | RNF40 |
| brl2 | E3 ubiquitin-protein ligase brl2 | HULC | RNF20, RNF40 |
| ccq1 | Structural maintenance of chromosomes protein ccq1 | Stn1-Ten1 | |
| clr4 | Histone H3-K9 methyltransferase | CLRC | SUV39H1, SUV39H2 |
| hta1 | Histone H2A-α | H2AFX | |
| mms19 | MMS19 nucleotide excision repair protein homolog | MMS19 | |
| raf1 | Rik1-associated factor 1 | CLRC | |
| rhp6 | Ubiquitin-conjugating enzyme E2 2 | HULC | UBE2B |
| rdp1 | RNA-dependent RNA polymerase 1 | ||
| shf1 | Small histone ubiquitination factor 1 | HULC | PPHLN1 |
| swi6 | Chromatin-associated protein swi6 | CBX1, CBX3, CBX5 | |
| tas3 | RNA-induced transcriptional silencing complex protein tas3 | RITS |
Annotation provides a schematically description of the gene product as indicated in UniprotKB database (http://www.uniprot.org/).
Potential human ortholog genes found in the databases PomBase or KOG (asterisks).
Histone acetylation is a dynamic process that affects the transcriptional state of chromatin and thus, transcriptionally active genes are usually enriched in hyperacetylated histones, whereas silent genes are associated to hypoacetylated histones.11 The HAT and HDAC complexes control the levels of histone acetylation by catalyzing the addition and the removal, respectively, of acetyl groups to lysine residues within histones. Both HATs and HDACs are classified into several families that are frequently conserved from yeast to humans.12 The HAT genes whose deletion contributes to 5FU cytotoxicity specifically encode for components of the SAGA and the nuA4 complexes, responsible for acetylating histone H313 and H4-H2A,14 respectively. Similarly, the absence of HDACs that form the Rpd3 complex15 also enhances the 5FU-induced cytotoxicity. The evolutionarily conserved Set1C/COMPASS complex that promotes transcriptional activation by histone H3 lysine 4 (H3K4) methylation16,17 seems to participate as well in 5FU response.
In addition to the post-translational modifications of histones, the replacement of the canonical histone H2A with the variant H2A.Z (encoded by pht1 in fission yeast) could be affected in 5FU-treated cells, because mutants lacking subunits of the Swr1 complex showed drug sensitivity. H2A.Z is highly conserved in evolution and it is important for cell cycle progression and for the establishment of active/inactive chromatin domains.18 Moreover, H2A.Z plays a role in centromere function and chromosome segregation in both S. pombe19 and mammals20 in which it is a structural component of centromeres.21
Interestingly, many strains deleted for heterochromatin components (HULC, RITS, CLRC and Stn1-Ten1 complexes) were sensitive to 5FU. The histone H2B ubiquitylation ligase complex (HULC) localizes to centromeric regions of S. pombe and human cells where it promotes transcriptional derepression.22,23 The Clr4 methyltransferase complex (CLRC) is responsible for histone H3 methylation on lysine 9 (H3K9me), which serves as a platform to recruit proteins required for nucleation and spreading of heterochromatin.24 CLRC was found to interact with the RITS (RNA-induced transcriptional silencing) complex, which functions as a physical bridge between noncoding RNA scaffolds and chromatin.25,26 The Stn1-Ten1 complex is critical for telomere protection in fission yeast27 and humans.28 Despite the fact that the collection tested only included the strain deleted for ccq1 but not stn1 or ten1, both genes are notably induced after 5FU treatment,6 suggesting that the length and silencing of chromosome ends might be affected.
The finding that mutants in these processes show a dramatic decrease in viability when exposed to 5FU, suggests that this drug might affect to some extent the post-translational modification of histones as well as the rate of histone exchange. Moreover, it also suggests that the combination of 5FU with drugs that modulate these epigenetic changes could enhance the effectiveness of the treatment. Interestingly, it has been previously reported that two histone deacetylase inhibitors (valproic acid and trichostatin A) synergistically potentiate the apoptotic effects of 5FU in human cancer cell lines.29,30 More recently, the overexpression of histone deacetylase 4 has been associated to 5FU-resistance in breast cancer cells,31 whereas strains deleted for histone acetyltransferase genes in the yeast Saccharomyces cerevisiae have been found to be sensitive to 5FU.32 Taken all this into account, it is possible that the inhibition of other epigenetic processes apart from histone deacetylation, such as acetylation, methylation or replacement of histones, could also improve the efficacy of 5FU-based treatments. Since modification of histones affecting heterochromatic regions and chromosome segregation might represent new intriguing targets for 5FU, we decided to focus on the effects caused by this drug in both processes.
Mutants defective in heterochromatin and chromosome segregation are sensitive to 5FU
The screening of mutants affecting 5FU sensitivity revealed a significant overrepresentation of strains deleted for genes involved in chromosome segregation, including those directly related to heterochromatin formation (Fig. 1). There are three main heterochromatic regions in fission yeast (centromeres, telomeres and the mating-type locus) that contain repetitive non-coding elements known as dg and dh repeats, whose bidirectional transcription generates double-stranded RNA (dsRNA). These molecules are rapidly processed into small interfering RNA (siRNA) to facilitate the recruitment of chromatin-modifying enzymes necessary for heterochromatin silencing. There is evidence suggesting that the mechanism by which interference RNA (RNAi) triggers heterochromatin assembly at centromeres could be conserved in mammals, especially in stem cells. For example, the deficiency of Dicer, which cleaves dsRNA to siRNA, leads to defective silencing and lower methylation levels of H3K9 at centromeres in mouse embryonic stem cells33 in addition to an aberrant accumulation of transcripts from centromeric repeats in a chicken–human hybrid cell line.34
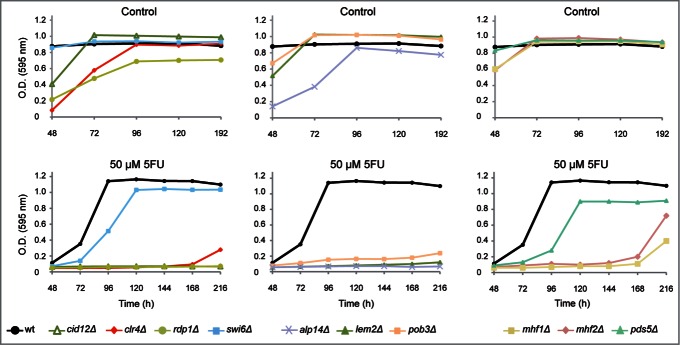
Interestingly, strains deleted for genes involved in RNA-interference-directed chromatin silencing such as cid12Δ, rdp1Δ and clr4Δ mutants were hypersensitive to 5FU (Fig. 2). Cid12 (a polyA polymerase) and Rdp1 (an RNA-directed RNA polymerase) are members of the RNA-directed RNA polymerase complex (RDRC), which is required for dsRNA synthesis and siRNA production.35 Clr4, a homolog of human SUV39h, is the only enzyme in S. pombe able to methylate H3K9, which represents a specific tag for transcriptionally silenced heterochromatin. The absence of H3K9me in cells depleted for Clr4/SUV39h impedes the recruitment of the chromodomain protein Swi624,36 causing heterochromatin defects and genome instability in fission yeast24,37 and mammals.38 Swi6 is closely related to the human heterochromatin protein 1 (HP1) and both promote the assembly and the spreading of heterochromatic structures.39,40 Moreover, Swi6 is also capable of recruiting cohesin to the dg and dh centromeric repeats ensuring a proper sister chromatid cohesion at centromeres.41 Pds5, a highly conserved protein, contributes to centromeric cohesion by stabilizing the cohesin complex.42-44 The absence of both Swi6 and Pds5 caused a reduction in the growth rates in cells treated with 5FU (Fig. 2). The effect of 5FU in the chromosome segregation process is further supported by the significant drug sensitivity of mutants involved in the kinetochore function (alp14Δ, mhf1Δ and mhf2Δ). The kinetochore is a multisubunit complex that assembles onto the centromeric region to mediate chromosome attachment to spindle microtubules. Mhf1 and Mhf2 play an important role in the assembly of the kinetochore structure,45 as do their corresponding human homologs CENP-S (Mhf1) and CENP-X (Mhf2). In fact, the CENP-S-X complex forms a unique chromatin structure similar to canonical nucleosomes at the centromeric regions.46 We also found that the fission yeast CKAP5 homolog Alp14, a protein that connects the mitotic spindles to the kinetochore47 and the nuclear membrane protein Lem2 (LEM2 in humans)48 contribute to the 5FU response. Similarly, the strain disrupted for Pob3, a component of the FACT complex that acts as a general chromatin factor reorganizing nucleosomes, showed 5FU hypersensitivity.
Figure 2.
Representative growth patterns of mutants deleted for chromosome segregation genes in the presence of 5FU. Growth analysis of the most sensitive chromosome segregation mutants to 50 μM 5FU compared with the untreated control. The growth of the strain swi6Δ and the control wild-type (ED668) are also shown. Data are representative of three independent experiments.
5FU decreases the transcript levels of the heterochromatic repeats
The significant 5FU sensitivity of strains deleted for genes involved in heterochromatin raises the possibility that the transcriptional silencing of the heterochromatic regions could be affected in cells exposed to the drug. As previously mentioned, the bidirectional expression of the dg and dh repeats by the RNA polymerase II has been shown to trigger siRNA production and heterochromatin formation in S. pombe, particularly at centromeres. The number and relative orientation of these repeats is different in the three fission yeast centromeres, corresponding the simplest organization to chromosome 1 that only contains one copy of dg and dh on both centromere flanks. In order to check whether the levels of these transcripts were affected by 5FU treatment, we examined the transcriptional profile of the centromeric regions that included data from the repetitive elements. The analysis revealed that the amount of transcripts detected for both sense and antisense heterochromatic repeats were notably lower after 5FU exposure (Fig. 3A). Strand-specific reverse transcription followed by real-time quantitative PCR (qPCR) experiments further confirmed a reduction in the levels of sense and antisense transcripts from dh (∼2-fold) and dg (∼3-fold) repeats compared to the untreated control (Fig. 3B). Interestingly, these results suggest that 5FU affects the transcriptional state of heterochromatin. An apparent paradox is that the transcription of heterochromatin is a key to its silencing, but heterochromatin actually functions as a dynamic structure whose expression is cell cycle regulated from yeast to mammalian cells.49,50 Despite its silent state throughout most of the cell cycle, the partial disruption of heterochromatin structure during S phase enables the transcription machinery to access these regions leading to the expression of centromeric repeats in S. pombe.51 Thus, the decreased levels of dg and dh transcripts might be a consequence of the delayed entry into S phase detected in 5FU-treated cells.6 This capacity of 5FU to block G1/S phase has also been reported in human cell lines.52 Given that the dg/dh transcripts trigger heterochromatin formation in S. pombe,53 it could be possible that 5FU affects the structure of heterochromatic regions. This possibility was further supported by the 5FU sensitivity of strains deleted for genes involved in RNAi-mediated heterochromatin formation (Fig. 2; Table S1). One of these strains, ago1Δ, showed an increased susceptibility to the drug (Fig. S2) but at lower levels than the established threshold for Table S1. The fission yeast protein Ago1 (homolog of human AGO1) directly binds to siRNA molecules and directs them to their complementary target, the centromeric transcripts. This interaction is proposed to recruit histone-modifying enzymes that favor chromatin silencing.25 Curiously, most of the siRNA associated with Ago1 contains a uracil at the 5´ position, a preference that has been attributed to higher stability of 5´U siRNA.54 Considering that 5FU could arise at that position instead of uracil, the binding properties and therefore the protein function could be affected in treated cells.
Figure 3.
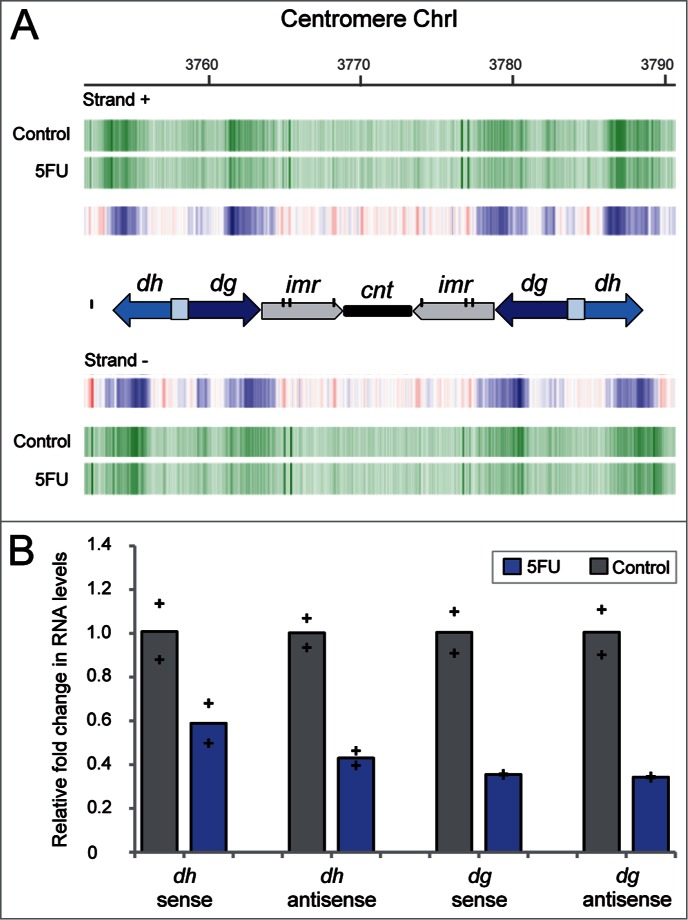
Decreased levels of centromeric dg and dh transcripts in fission yeast cells exposed to 5FU. (A) Expression profile of centromere 1 regions obtained by microarray analysis. Vertical green lines represent transcription from sense (+) and antisense (−) DNA strand in cells exposed to 5FU for 60 min or untreated control. Blue vertical lines indicate differential under-represented transcripts between both conditions. The diagram shows the organizational structure of centromere 1: the dg/dh repeats (whose relative orientation is indicated by arrows), imr (inner repeats), cnt (central core domain) and individual copies of tRNA genes that are represented by vertical black lines. Note that the dg and dh elements are found in multiple copies at heterochromatic regions and therefore, the amount of transcripts detected are representative of the expression levels of the individual repetitions. The downregulation of dg/dh transcripts was also observed after 240 min of drug treatment (data available through the genome browser). (B) Relative fold change in transcripts abundance between untreated and 5FU-treated cells determined by strand-specific reverse transcription followed by qPCR. Bars represent the average data for two independent biological replicates with each of the individual data points being displayed by a cross. Two-tailed Student's t test revealed that differences were statistically significant in all cases (P value < 0 .05) except for transcript dh sense (P = 0.11).
5FU reduces histone H3 lysine 9 dimethylation levels relative to total H3
The diminished levels of centromeric transcripts detected in cells treated with 5FU (Fig. 3) could cause a defect in the normal structure of heterochromatin. To test this, we compared the levels of methylation of histone H3 at lysine 9, a conserved hallmark of heterochromatin, as well as total H3, for selected heterochromatic regions in cells exposed to 5FU with respect to untreated controls using chromatin immunoprecipitation (ChIP) followed by qPCR experiments. The H3K9me2 and total H3 profiles detected in untreated cells (Fig. 4A) coincided with previous data55,56 showing preferential enrichment at centromeric repeats and a notable decrease over the tRNA genes that marked the boundaries of heterochromatic dg/dh regions with the surrounding euchromatic regions or the central domain. The innermost repeats (imr) and the central core (cnt) form the central domain, which contains the histone H3 variant Cnp1 (CENP-A). Our results revealed that cells treated with 5FU showed a slight decrease in the absolute levels of H3K9me2 at the centromeric heterochromatin (Fig. 4B), whereas a significant ∼1.7-fold increase in total H3 (P value < 0.05) was detected (Fig. 4C). The higher level of H3 suggests an increment in the nucleosome occupancy state at centromeres that might act as a physical barrier for the transcriptional machinery contributing to dg/dh repression. This effect seemed to be specific to these regions because the genome-wide nucleosome density obtained by chromatin digestion with micrococcal nuclease (MNAse) did not significantly changed in cells treated with 5FU compared to the control (Fig. S3). To determine the relative enrichment of H3K9me2 we normalized to total H3 and thus, we found a ∼2-fold decrease in the ratio H3K9me2/H3 in 5FU-treated cells (Fig. 4D). This reduction could be a consequence of the low levels of the centromeric transcripts in cells treated with 5FU (Fig. 3), since they are required to methylate H3K9. Regardless of the specific mechanism, the decreased H3K9me2 levels may contribute to 5FU toxicity since mutants defective in H3K9 methylation such as clr4Δ,57 rdp1Δ,58 tas3Δ59 and lsd1Δ60 are hypersensitive to the drug (Table S1). Interestingly, the overexpression of tRNA genes detected in cells treated with 5FU6 might be important in preventing the spread of H3K9me2 beyond the heterochromatin boundaries, since the association of centromeric tRNA genes with RNA polymerase III complex is required for barrier activity.61 It is also interesting to note the 5FU-sensitivity of the strain deleted for fft3, which mediates protection from euchromatin formation in the central core region.56
Figure 4.
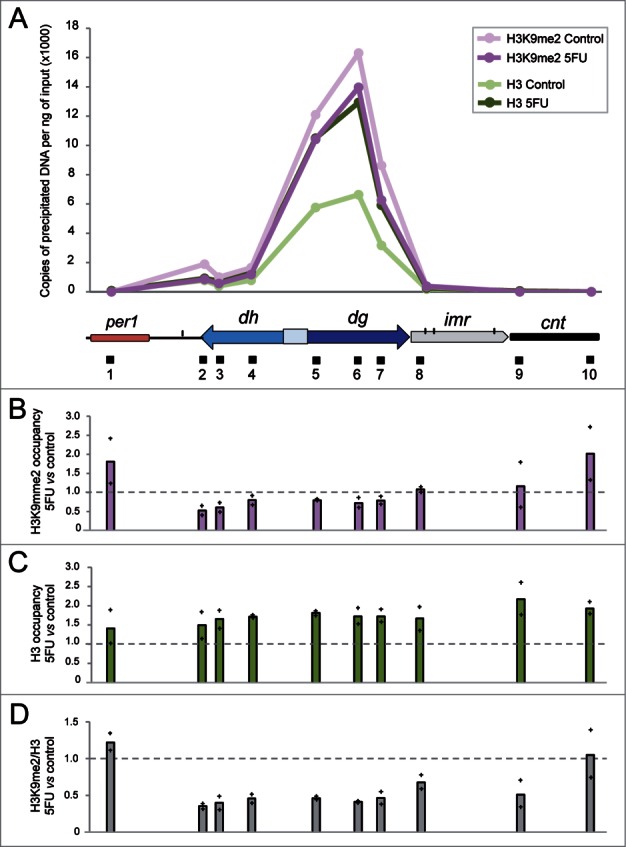
5FU diminishes H3K9me2 levels relative to total H3. (A) Representative distribution of H3K9me2 and H3 at centromere 1 in S. pombe cells treated or not with 5FU. The values represent the number of DNA copies per ng of input obtained by ChIP experiments followed by qPCR analysis. The diagram below the graph shows the organization of a part of centromere 1 in addition to per1, the euchromatic gene closest to the left dg/dh repeats. Specific regions amplified by qPCR are indicated by black boxes beneath the diagram. Note that since the dg and dh elements are repetitive elements, their relative histone enrichment is a mixture of the individual repetitions located at the three centromeres. However, the other amplification regions were located in the unique genomic position indicated. (B) H3K9me2 occupancy in 5FU-treated cells compared to the untreated control. (C) H3 occupancy in 5FU-treated cells compared to the untreated control. (D) Relative H3K9me2/H3 enrichment in cells exposed to 5FU with respect to the untreated control. Bars represent the average data for two independent replicates with each of the individual data points being displayed by a cross.
Epigenetic alterations that influence chromatin accessibility and gene expression have been found in many tumor cell types.62,63 However, the potential effects of anticancer drugs in histone modifications remain largely unexplored. Intriguingly, the histone modification levels have been shown to predict response to 5FU chemotherapy in patients with pancreatic cancer. In particular, low levels of H3K9me2 were significantly associated with poor survival outcomes after 5FU treatment.64 Given that the presence of the epigenetic mark H3K9me2 is necessary to recruit chromatin-modifying factors involved in the maintenance and spreading of heterochromatin,65 it is conceivable that 5FU-treated cells may show deficiencies in processes, such as chromosomal segregation, that required an intact heterochromatin structure at the centromeres.
5FU causes defects in chromosome segregation
Exploring the mode of action of 5FU by screening a haploid deletion library could underestimate the role of chromosome segregation genes, because most of them are essential for yeast viability. An alternative approach to identifying the genes implicated in the 5FU response involves examining the transcriptional changes associated with drug exposure. This analysis revealed that genes involved in chromosome segregation (N = 166) showed significantly higher differential expression levels than those included in other categories (P value < 0.0001) (Fig. 5A). Chromosome segregation genes with induction levels above the genomic threshold (0.06) encode for proteins that belong to three main complexes: Mis6-Sm4, cohesin and NMS (Fig. 5B). The Mis6-Sim4 complex mediates the loading of the histone H3 variant Cnp1/CENP-A onto centromeres leading to kinetochore assembly and mitotic progression from fission yeast to mammals.66 Mitotic cohesin is a multiprotein complex required for sister chromatid cohesion, spindle pole formation and chromosome movement in eukaryotes.67 On the other hand, the NMS complex is necessary for the attachment of spindle microtubules to the kinetochore.68 The up-regulation of these genes probably represents a cellular attempt to counteract drug damage providing further evidence of the effect of 5FU on chromosome segregation. The 5FU hypersensitivity of the only two viable mutants deleted for genes represented in Fig. 5B (pds5Δ and dad1Δ; Table S1) further support this possibility. To check whether 5FU produces any defect in chromosome segregation in S. pombe, we monitored chromosome loss events using a strain carrying an additional and non-essential chromosome called Ch16. Ch16 is a highly stable 530 Kb linear mini-chromosome69 that includes the adenine allele ade6-M216. Strains carrying Ch16 form white colonies on plates containing a limited amount of adenine due to intragenic complementation between ade6-M216 and ade6-M210 on chromosome III. In contrast, loss of Ch16 results in a red colony or sector because of the accumulation of a red adenine precursor. Interestingly, we found that 5FU increased the rate of mini-chromosome Ch16 loss by more than 18-fold compared to the untreated control (Fig. 5C). This effect could be a consequence of an altering mitotic spindle function, which occurs in numerous heterochromatin mutants.70 Consistent with this hypothesis, we observed that thiabendazole (TBZ), a microtubule-destabilizing drug, potentiates the cytotoxic effects of 5FU (Fig. 5D). Furthermore, this drug combination could be potentially used in cancer therapy since TBZ has been recently shown to be a promising candidate in the treatment of solid tumors because it slows tumor growth as it rapidly depolymerizes microtubules of the newly formed vasculature.71 Similarly, other microtubule-targeting drugs such as cryptophycins, docetaxel and epothilone D synergistically increased the apoptotic effect of 5FU in human tumor xenografts,72 murine tumor models73 or human cancer cell lines.74
Figure 5.
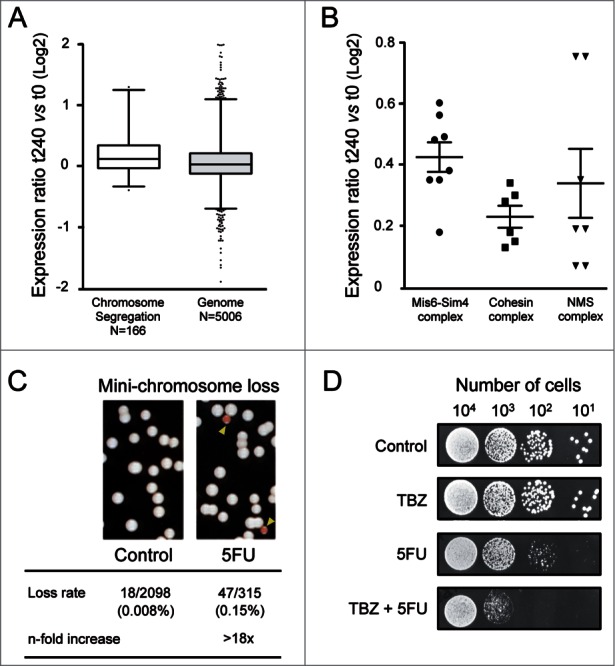
Chromosomal segregation defects produced by 5FU. (A) Box and whisker plots showing the log2-fold changes in gene expression (measured by microarray probe intensities) after 240 min of 5FU treatment relative to untreated control (t240 vs t0) for each category. The individual boxes represent the median (central horizontal line) and the 75–25% percentiles. The whiskers extend from the boxes to 1% and 99% of the data set. Dots indicate outliers. Data are representative of two independent experiments. Two-tailed Mann-Whitney test revealed that differences were statistically significant (P value < 0.0001). The values of the twelve genes with probe intensity levels above or below two were omitted for clarity. (B) The log2-fold differential expression ratios (5FU 240 min compared to control) for individual chromosome segregation genes with higher induction levels than the genomic threshold (0.06). Genes encode for subunits of three main complexes Mis6-Sim4, cohesin and NMS. Horizontal lines represent mean values and vertical lines the standard error of mean. (C) Rate of the mini-chromosome Ch16 loss (monitored by the appearance of red colonies/sectors on low adenine plates) was >18-fold higher in cells exposed to 5FU. (D) The combined treatment of 5FU with the microtubule-destabilizing agent thiabendazole (TBZ) synergistically enhanced the cytotoxic effects of 5FU. Serial dilutions of S. pombe cells showing the cellular sensitivity to 5FU (150 μM), TBZ (20 μM) or both combined.
Together, these results suggest that 5FU in S. pombe affects the segregation of chromosomes, a process that depends to a large extent on the presence of an intact heterochromatin structure at the centromeres. It might be possible that 5FU produces similar defects in human cells, since the structural and functional properties of centromeres are conserved from fission yeast to humans.75 In both cases centromeres are composed of repetitive elements (α satellite in human, dg/dh elements in S. pombe) that are marked by epigenetic methylation of H3K9 and associated to evolutionary conserved proteins involved in heterochromatin formation and/or centromeric function. In addition, heterochromatin silencing in mammals is also closely intertwined with non-coding RNAs, which provide the initial scaffolding needed in recruiting chromatin-modifying complexes that regulate transcription.76,77 Thus, the incorporation of 5FU into these non-coding RNAs could affect the transcriptional state and the structure of heterochromatin in human cells as it occurs in fission yeast.
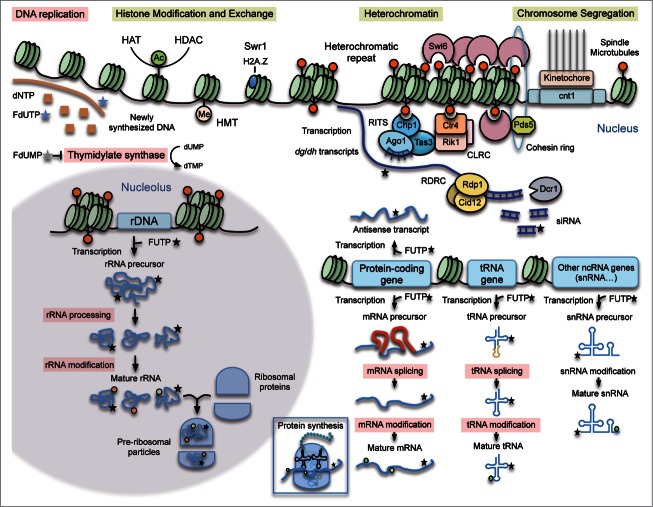
Previous studies have shown that the impairment of the RNA metabolism is responsible to a large extent for the cytotoxic effects of 5FU.1 Here, we provide evidence showing that 5FU also interferes with chromosome organization (nucleosome occupancy, histone modification/exchange, heterochromatin silencing) and segregation in eukaryotes (Fig. 6). Since many of these cellular processes are conserved in human cells, these findings point to particular new targets and drug combinations that could increase the effectiveness of 5FU-based treatments.
Figure 6.
Mechanism of 5FU action. Summary of the major 5FU targets previously described in eukaryotic cells (pink boxes) and new potential ones predicted by our analysis in the fission yeast model (yellow boxes). 5FU is converted intracellularly into three main active metabolites: fluorodeoxyuridine monophosphate (FdUMP), fluorodeoxyuridine triphosphate (FdUTP) and fluorouridine triphosphate (FUTP). FdUMP affects DNA replication because it inhibits the thymidylate synthase causing a decrease in the production of dTMP. FdUTP misincorporated into DNA during replication interferes with the normal DNA metabolism. FUTP can be misincorporated into different types of RNA molecules directly involved in protein synthesis (mRNA, tRNA, rRNA, snRNA) or in other processes such as RNA-mediated heterochromatin formation (dg/dh transcripts) or transcription regulation (antisense transcripts). These 5FU-containing RNAs can undergo defective processing and post-transcriptional modifications affecting their normal functions. The new proposed 5FU targets in eukaryotic cells include heterochromatin, chromosome segregation and histone modification/exchange. When incorporated into centromeric transcripts, FUTP could alter the RNA-mediated heterochromatin formation. In S. pombe the RNA-directed RNA polymerase complex (RDRC) copies the centromeric transcripts into dsRNA that are further processed by Dicer (Dcr1) into siRNA. The RITS complex loads these siRNA molecules, which facilitates its positioning at heterochromatic regions. The Clr4 methyltransferase complex (CLRC) methylates H3K9 providing a platform to recruit proteins necessary for heterochromatin formation and spreading. Chromosome segregation is also affected in 5FU-treated cells maybe as an indirect consequence of heterochromatic defects.
Materials and Methods
Chemicals
5FU was supplied by Sigma-Aldrich (F6627) prepared as a 20 mM stock solution in water and kept at 4 °C. Thiabendazole (TBZ), obtained from Sigma-Aldrich (T5535), was dissolved in DMSO as a stock solution at 20 mg/ml and stored at room temperature.
Genome-wide screen of deletion strains sensitive to 5FU
The S. pombe haploid deletion library used in this study was obtained from Bioneer (Mutant Sets ver 2.0 and 3.0) (http://pombe.bioneer.co.kr/) and contains 3,310 single-gene deletion mutants that represent approximately 90% of the non-essential genes in fission yeast. The 5FU-sensitivity was analyzed three times for each individual deletion strain of the Bioneer collection. Strains were inoculated from frozen stocks into 96-well plates containing YE medium (3% glucose, 0.5% yeast extract) and grown to saturation at 30°C. Cultures were then replicated onto 96-well plates with YE medium supplemented with different concentrations of 5FU (0, 25, 50 and 150 μM) using a stainless steel 96-pin replicator (Nalgene Nunc International) and incubated at 30°C. Growth was quantitatively scored every 24 h by monitoring the absorbance at 595 nm with a microplate reader (Varioskan, Thermo Scientific). A total of 270 deletion strains with enhanced sensitivity to 5FU compared to the wild-type were selected based on their growth inhibition at 150 μM 5FU (Table S1). Mutants that showed a reduced growth in the YE control were not included in further analysis as well as those disrupted for genes either with an unknown function or implicated in specific yeast pathways. We estimated by PCR experiments that approximately 90% of the strains had the correct target gene deleted according to Bioneer database (Fig. S4). Primers for amplification reactions are listed in Table S2.
Gene Ontology classification and interaction network analysis
Genes whose disruption confers sensitivity to 5FU were classified according to the GO database into biological processes. The statistical significance of the groups was calculated using the two-sided Fisher´s exact test. The protein interaction network was generated using the STRING v9.05 database (http://string905.embl.de) and their default parameters including the confidence score >0.4 were applied. The significance of the GO cellular component terms was also evaluated with STRING.
Strand-specific reverse transcriptase and real-time quantitative PCR analysis
To determine the levels of sense and antisense transcripts from dg and dh elements, we first synthesized DNA products from total RNA as follows: 200 ng of total RNA was reverse transcribed with AMV Reverse Transcriptase (New England Biolabs, M0277) with a specific primer (Table S2) according to the manufacturer's recommendations. Samples omitting reverse transcriptase were included as negative controls in each set of reactions. Quantification was performed by qPCR in triplicates on a BioRad CFX 384 instrument with 10 μl of final reaction volume containing 5 ng of DNA, 2.5 μM of each forward and reverse primer and 5 μl of Power Sybr Green PCR Master Mix (Applied Biosystems, PN 4367659). The qPCR assays were set up using an Eppendorf pipetting robot (epMotion 5075). Cycling parameters were as follows: 10 sec at 95°C followed by 40 2-step cycles of 95°C for 15 sec + 60°C for 60 sec. Melting curve analysis from 60°C to 95°C was included at the end of the program. Data analysis was performed using GenEx qPCR data analysis software v.5.3.7 (MultiD). Primers for amplification reactions are listed in Table S2.
Chromatin immunoprecipitation
Cultures of S. pombe wild-type strain 972 h− were grown in YE medium at 30°C until the early exponential phase (OD595 = 0.2, ∼4 × 106 cells/ml). 5FU was added to the cultures except for the controls to a final concentration of 500 μM and incubation was allowed to proceed for 240 min. Two biological replicate experiments from independent cell cultures were performed. Cells were cross-linked with 1% formaldehyde for 30 min at room temperature and then, the reaction was stopped by adding glycine to a final concentration of 125 mM. All the following steps were carried out at 4°C unless otherwise stated. The pellets were washed twice with TBS (50 mM Tris-HCl, pH 7.5, 150 mM NaCl) and resuspended in 0.4 ml of lysis buffer (50 mM Hepes-KOH, pH 7.5, 140 mM NaCl, 1mM EDTA, 1% (v/v) Triton X-100, 0.1% (w/v) sodium deoxycholate) supplemented with protease inhibitors (Complete Mini EDTA-free, Roche) according to the manufacturer's instructions. The cells were disrupted with glass beads using a FastPrep-24 (MP Biomedicals) and the lysates were sonicated in a Bioruptor (Diagenode) for 35 cycles of 30 sec at high power and 30 sec off, yielding chromatin fragments of approximately 300 bp average size. The sonicated material was centrifuged at 13000 g for 30 min to remove cell debris. An aliquot of 50 μl of soluble chromatin was kept to determine the amount of input DNA, while the rest was pre-cleared to reduce non-specific background with 80 μl 50% (v/v) Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, sc-2003) by shaking in a Labinco rotator during 1 h. After centrifugation at 500 g for 1 min, the same amount of pre-cleared chromatin (200 μl) was immunoprecipitated with either 4 μg of anti-H3K9m2 antibody (Abcam, ab1220) or 4 μg of anti-H3 antibody (Abcam, ab1791). An aliquot of 50 μl with no antibody was used as a non-immunoprecipitated control. Overnight incubation was followed by the addition of 80 μl 50% (v/v) Protein A/G PLUS-Agarose to each sample, after which the samples were incubated with rotation for a further 2 h. Beads were washed 3 times in each of these buffers: RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40), LiCl buffer (10 mM Tris-HCl, pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA) and TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA). The antibody-protein-DNA complexes were eluted by incubating the beads twice with 0.25 ml elution buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 1% SDS) for 5 min at 65°C and then 15 min at room temperature with rotation. To reverse the cross-links the samples were incubated overnight at 65°C after the addition of NaCl to a final concentration of 0.2 M. Samples were then treated with 2 μl DNAse-free RNAse (0.5 μg/μl) for 1 h and then with 5 μl proteinase K (10 μg/μl) during 3 hours at 42°C. DNA was purified by phenol-chloroform extraction followed by ethanol precipitation and then resuspended in 120 μl TE buffer. Absolute quantification of DNA derived from ChIP analysis was essentially performed as previously described for dg and dh elements except for some modifications. Briefly, reactions were carried out in 10 μl of final volume containing 1 μl of the samples, 2.5 μM of each forward and reverse primer and 5 μl of SsoFast EvaGreen SuperMix (Bio-Rad, 172–5204). Cycling conditions were 5 sec at 95°C followed by 40 2-step cycles of 95°C for 5 sec + 62°C for 5 sec. Primers for amplification reactions are listed in Table S2.
Mitotic chromosome stability assays
Cells of S. pombe strain h− ade6-M210 Ch16 (ade6-M216) growing exponentially (OD595 = 0.4, ∼8 × 106 cells/ml) in YE medium were plated in YE plates containing or not 500 μM 5FU and were incubated at 32°C for 15 d followed by 2 d at 4°C to allow for a deep red color to appear. Colonies with red sectors were counted. To calculate the rate of mini-chromosome Ch16 loss, the total number of colonies were divided by the number of red-sectored colonies.
Chromatin digestion with micrococcal nuclease
Chromatin isolation for micrococcal nuclease digestion was essentially performed as previously described78 except for some modifications. Briefly, 5FU was added to 100 ml of early exponentially growing S. pombe wild-type 972 h− cells (OD595 = 0.2, ∼4 × 106 cells/ml) to a final concentration of 500 μM, except for the controls. After 240 min at 30°C, ∼5 × 108 cells from cultures treated or not with 5FU were harvested, washed once with cold water and incubated under shaking during 10 min in 10 ml of preincubation solution (20 mM citric acid, 20 mM Na2HPO4, 40 mM EDTA, pH 8.0, 30 mM β-mercaptoethanol). Then, the cells were incubated in 10 ml of Sorbitol/Tris buffer (1 M sorbitol, 50 mM Tris-HCl, pH 7.4, 10 mM β-mercaptoethanol) with 30 mg zymolyase 20T for 30 min under shaking. The resulting spheroplasts were washed once with Sorbitol/Tris buffer without β-mercaptoethanol, resuspended to a final volume of 3.2 ml of NP-buffer (1 M sorbitol, 50 mM NaCl, 10 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 1 mM CaCl2, 0.75% NP-40, 1 mM β-mercaptoethanol and 0.5 mM spermidine) and divided into 4 aliquots of 0.5 ml. MNase (New England Biolabs, M0247S) was added to each sample to a final concentration of 0, 15, 60 and 240 U/ml and incubation was allowed to proceed for 10 min at 37 ºC, after which the reaction was stopped by the addition of 65 μl of 5% SDS/100 mM EDTA, pH 8.0. Samples were incubated overnight at 37°C with 10 μl of Proteinase K (stock solution 10 μg/μl), and then with 1 μl DNase-free RNase (0.5 μg/μl) during 1 h. DNA was purified by phenol-chloroform extraction followed by ethanol precipitation, and then resuspended in 50 μl TE buffer.
Data access
The transcriptional profile of cells treated with the drug was reprocessed to include the repetitive elements that were filtered out in our previous work.6 Hybridization signals from probes that map to more than one genomic localization were normalized relative to the number of repeats. The subsequent transcriptional genome-wide response induced by 5FU in S. pombe can be accessed from a searchable genome browser at http://genomics.usal.es/cgi-bin/gb2/gbrowse/sp_moj3/.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Dr. Luis Quintales for reprocessed microarray data to include repetitive elements. We thank Dr. María Gómez and Dr. Crisanto Gutiérrez for providing antibodies, Dr. Joana Sequeira-Mendes for advice with ChIP experiments and the Massive Sequencing service (CBMSO, Madrid) for assistance in qPCR experiments.
Funding
This work was supported by the Spanish Ministry of Economy and Competitiveness (BFU2011–23645) to MS, the CONSOLIDER-INGENIO from the Spanish Ministry of Science and Innovation CSD2007–00015 to MS, the Fundación Mutua Madrileña to MS and an Institutional Grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.” LM was holder of a CONSOLIDER-INGENIO contract.
References
- 1. Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 2003; 3:330-8; PMID:12724731; http://dx.doi.org/ 10.1038/nrc1074 [DOI] [PubMed] [Google Scholar]
- 2. Santi DV, McHenry CS, Sommer H. Mechanism of interaction of thymidylate synthetase with 5-fluorodeoxyuridylate. Biochemistry 1974; 13:471-81; PMID:4203910; http://dx.doi.org/ 10.1021/bi00700a012 [DOI] [PubMed] [Google Scholar]
- 3. Parker WB, Cheng YC. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol Ther 1990; 48:381-95; PMID:1707544; http://dx.doi.org/ 10.1016/0163-7258(90)90056-8 [DOI] [PubMed] [Google Scholar]
- 4. Kufe DW, Major PP. 5-Fluorouracil incorporation into human breast carcinoma RNA correlates with cytotoxicity. J Biol Chem 1981; 256:9802-5; PMID:7275977 [PubMed] [Google Scholar]
- 5. Glazer RI, Lloyd LS. Association of cell lethality with incorporation of 5-fluorouracil and 5-fluorouridine into nuclear RNA in human colon carcinoma cells in culture. Mol Pharmacol 1982; 21:468-73; PMID:7099147 [PubMed] [Google Scholar]
- 6. Mojardin L, Botet J, Quintales L, Moreno S, Salas M. New insights into the RNA-based mechanism of action of the anticancer drug 5’-Fluorouracil in eukaryotic cells. PLoS One 2013; 8:e78172; PMID:24223771; http://dx.doi.org/ 10.1371/journal.pone.0078172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takeda K, Mori A, Yanagida M. Identification of genes affecting the toxicity of anti-cancer drug bortezomib by genome-wide screening in S. pombe. PLoS One 2011; 6:e22021; PMID:21760946; http://dx.doi.org/ 10.1371/journal.pone.0022021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou X, Ma Y, Fang Y, gerile W, Jaiseng W, Yamada Y, Kuno T. A genome-wide screening of potential target genes to enhance the antifungal activity of micafungin in Schizosaccharomyces pombe. PLoS One 2013; 8:e65904; PMID:23738021; http://dx.doi.org/ 10.1371/journal.pone.0065904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fang Y, Hu L, Zhou X, Jaiseng W, Zhang B, Takami T, Kuno T. (2012) A genomewide screen in Schizosaccharomyces pombe for genes affecting the sensitivity of antifungal drugs that target ergosterol biosynthesis. Antimicrob Agents Chemother 2012; 56:1949-59; PMID:22252817; http://dx.doi.org/ 10.1128/AAC.05126-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang L, Ma N, Liu Q, Ma Y. Genome-wide screening for genes associated with valproic acid sensitivity in fission yeast. PLoS One 2013; 8:e68738; PMID:23861937; http://dx.doi.org/ 10.1371/journal.pone.0068738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell 2006; 23:289-96; PMID:16885019; http://dx.doi.org/ 10.1016/j.molcel.2006.06.017 [DOI] [PubMed] [Google Scholar]
- 12. Carrozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet 2003; 19:321-9; PMID:12801725; http://dx.doi.org/ 10.1016/S0168-9525(03)00115-X [DOI] [PubMed] [Google Scholar]
- 13. Rodriguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep 2009; 10:843-50; PMID:19609321; http://dx.doi.org/ 10.1038/embor.2009.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol 2004; 24:1884-96; PMID:14966270; http://dx.doi.org/ 10.1128/MCB.24.5.1884-1896.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol 2008; 9:206-18; PMID:18292778; http://dx.doi.org/ 10.1038/nrm2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dehé PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, Rodríguez-Gil A, Mkandawire M, Landsberg K, Shevchenko A, et al. . Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem 2006; 281:35404-12; http://dx.doi.org/ 10.1074/jbc.M603099200 [DOI] [PubMed] [Google Scholar]
- 17. Dehé PM, Géli V. The multiple faces of Set1. Biochem Cell Biol 2006; 84:536-48; http://dx.doi.org/ 10.1139/o06-081 [DOI] [PubMed] [Google Scholar]
- 18. Guillemette B, Gaudreau L. Reuniting the contrasting functions of H2A.Z. Biochem Cell Biol 2006; 84:528-35; PMID:16936825; http://dx.doi.org/ 10.1139/o06-077 [DOI] [PubMed] [Google Scholar]
- 19. Hou H, Wang Y, Kallgren SP, Thompson J, Yates JR, 3rd, Jia S. Histone variant H2A.Z regulates centromere silencing and chromosome segregation in fission yeast. J Biol Chem 2010; 285:1909-18; PMID:19910462; http://dx.doi.org/ 10.1074/jbc.M109.058487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rangasamy D, Greaves I, Tremethick DJ. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat Struct Mol Biol 2004; 11: 650-65; PMID:15195148; http://dx.doi.org/ 10.1038/nsmb786 [DOI] [PubMed] [Google Scholar]
- 21. Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ. H2A.Z contributes to the unique 3D structure of the centromere. Proc Natl Acad Sci U S A 2007; 104:525-30; PMID:17194760; http://dx.doi.org/ 10.1073/pnas.0607870104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zofall M, Grewal SI. HULC, a histone H2B ubiquitinating complex, modulates heterochromatin independent of histone methylation in fission yeast. J Biol Chem 2007; 282:14065-72; PMID:17363370; http://dx.doi.org/ 10.1074/jbc.M700292200 [DOI] [PubMed] [Google Scholar]
- 23. Sadeghi L, Siggens L, Svensson JP, Ekwall K. Centromeric histone H2B monoubiquitination promotes noncoding transcription and chromatin integrity. Nat Struct Mol Biol 2014; 21:236-43; PMID:24531659; http://dx.doi.org/ 10.1038/nsmb.2776 [DOI] [PubMed] [Google Scholar]
- 24. Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 2008; 15:381-8; PMID:18345014; http://dx.doi.org/ 10.1038/nsmb.1406 [DOI] [PubMed] [Google Scholar]
- 25. Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 2004; 303:672-6; PMID:14704433; http://dx.doi.org/ 10.1126/science.1093686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Creamer KM, Partridge JF. RITS-connecting transcription, RNA interference, and heterochromatin assembly in fission yeast. Wiley Interdiscip Rev 2011; RNA 2:632-46; http://dx.doi.org/ 10.1002/wrna.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin V, Du LL, Rozenzhak S, Russell P. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci U S A 2007; 104:14038-43; PMID:17715303; http://dx.doi.org/ 10.1073/pnas.0705497104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bryan C, Rice C, Harkisheimer M, Schultz DC, Skordalakes E. Structure of the human telomeric Stn1-Ten1 capping complex. PLoS One 2013; 8:e66756; PMID:23826127; http://dx.doi.org/ 10.1371/journal.pone.0066756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee JH, Park JH, Jung Y, Kim JH, Jong HS, Kim TY, Bang YJ. Histone deacetylase inhibitor enhances 5-fluorouracil cytotoxicity by down-regulating thymidylate synthase in human cancer cells. Mol Cancer Ther 2006; 5:3085-95; PMID:17172411; http://dx.doi.org/ 10.1158/1535-7163.MCT-06-0419 [DOI] [PubMed] [Google Scholar]
- 30. Iwahashi S, Ishibashi H, Utsunomiya T, Morine Y, Ochir TL, Hanaoka J, Mori H, Ikemoto T, Imura S, Shimada M. Effect of histone deacetylase inhibitor in combination with 5-fluorouracil on pancreas cancer and cholangiocarcinoma cell lines. J Med Invest 2011; 58:106-9; PMID:21372494; http://dx.doi.org/ 10.2152/jmi.58.106 [DOI] [PubMed] [Google Scholar]
- 31. Yu SL, Lee DC, Son JW, Park CG, Lee HY, Kang J. Histone deacetylase 4 mediates SMAD family member 4 deacetylation and induces 5-fluorouracil resistance in breast cancer cells. Oncol Rep 2013; 30:1293-300; PMID:23817620 [DOI] [PubMed] [Google Scholar]
- 32. Matuo R, Sousa FG, Bonatto D, Mielniczki-Pereira AA, Saffi J, Soares DG, Escargueil AE, Larsen AK, Henriques JA. ATP-dependent chromatin remodeling and histone acetyltransferases in 5-FU cytotoxicity in Saccharomyces cerevisiae. Genet Mol Res 2013; 12:1440-56; PMID:23661467; http://dx.doi.org/ 10.4238/2013.April.26.6 [DOI] [PubMed] [Google Scholar]
- 33. Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 2005; 19:489-501; PMID:15713842; http://dx.doi.org/ 10.1101/gad.1248505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol 2004; 6:784-91; PMID:15247924; http://dx.doi.org/ 10.1038/ncb1155 [DOI] [PubMed] [Google Scholar]
- 35. Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 2004; 119:789-802; PMID:15607976; http://dx.doi.org/ 10.1016/j.cell.2004.11.034 [DOI] [PubMed] [Google Scholar]
- 36. Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, Allshire R. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 1995; 269:1429-31; PMID:7660126; http://dx.doi.org/ 10.1126/science.7660126 [DOI] [PubMed] [Google Scholar]
- 37. Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev 1995; 9:218-33; PMID:7851795; http://dx.doi.org/ 10.1101/gad.9.2.218 [DOI] [PubMed] [Google Scholar]
- 38. Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. . Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 2001; 107:323-37; PMID:11701123; http://dx.doi.org/ 10.1016/S0092-8674(01)00542-6 [DOI] [PubMed] [Google Scholar]
- 39. Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 2001; 410:120-4; PMID:11242054; http://dx.doi.org/ 10.1038/35065138 [DOI] [PubMed] [Google Scholar]
- 40. Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 2003; 299:721-5; PMID:12560555; http://dx.doi.org/ 10.1126/science.1078572 [DOI] [PubMed] [Google Scholar]
- 41. Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science 2001; 294:2539-42; PMID:11598266; http://dx.doi.org/ 10.1126/science.1064027 [DOI] [PubMed] [Google Scholar]
- 42. Tanaka K, Hao Z, Kai M, Okayama H. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J 2001; 20:5779-90; PMID:11598020; http://dx.doi.org/ 10.1093/emboj/20.20.5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang SW, Read RL, Norbury CJ. Fission yeast Pds5 is required for accurate chromosome segregation and for survival after DNA damage or metaphase arrest. J Cell Sci 2002; 115:587-98; PMID:11861765 [DOI] [PubMed] [Google Scholar]
- 44. Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci 2005; 118:2133-41; PMID:15855230; http://dx.doi.org/ 10.1242/jcs.02355 [DOI] [PubMed] [Google Scholar]
- 45. Bhattacharjee S, Osman F, Feeney L, Lorenz A, Bryer C, Whitby MC. MHF1-2/CENP-S-X performs distinct roles in centromere metabolism and genetic recombination. Open Biol 2013; 3:130102; PMID:24026537; http://dx.doi.org/ 10.1098/rsob.130102 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46. Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 2012; 148:487-501; PMID:22304917; http://dx.doi.org/ 10.1016/j.cell.2011.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garcia MA, Vardy L, Koonrugsa N, Toda T. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J 2001; 20:3389-401; PMID:11432827; http://dx.doi.org/ 10.1093/emboj/20.13.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. King MC, Drivas TG, Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell 2008; 134:427-38; PMID:18692466; http://dx.doi.org/ 10.1016/j.cell.2008.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 2008; 451:734-7; PMID:18216783; http://dx.doi.org/ 10.1038/nature06561 [DOI] [PubMed] [Google Scholar]
- 50. Lu J, Gilbert DM. Cell cycle regulated transcription of heterochromatin in mammals vs. fission yeast: functional conservation or coincidence? Cell Cycle 2008; 7:1907-10; PMID:18604169; http://dx.doi.org/ 10.4161/cc.7.13.6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol 2008; 18:490-5; PMID:18394897; http://dx.doi.org/ 10.1016/j.cub.2008.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li MH, Ito D, Sanada M, Odani T, Hatori M, Iwase M, Nagumo M. Effect of 5-fluorouracil on G1 phase cell cycle regulation in oral cancer cell lines. Oral Oncol 2004; 40:63-70; PMID:14662417; http://dx.doi.org/ 10.1016/S1368-8375(03)00136-2 [DOI] [PubMed] [Google Scholar]
- 53. Aygün O, Grewal SI. Assembly and functions of heterochromatin in the fission yeast genome. Cold Spring Harb Symp Quant Biol 2010; 75:259-67; PMID:21502415; http://dx.doi.org/ 10.1101/sqb.2010.75.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bühler M, Spies N, Bartel DP, Moazed D. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol 2008; 15:1015-23; PMID:18776903; http://dx.doi.org/ 10.1038/nsmb.1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 2005; 37:809-19; PMID:15976807; http://dx.doi.org/ 10.1038/ng1602 [DOI] [PubMed] [Google Scholar]
- 56. Stråfors A, Walfridsson J, Bhuiyan H, Ekwall K. The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLoS Genet 2011; 7:e1001334; http://dx.doi.org/ 10.1371/journal.pgen.1001334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 2005; 309:467-9; PMID:15947136; http://dx.doi.org/ 10.1126/science.1114955 [DOI] [PubMed] [Google Scholar]
- 58. Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 2002; 297:1833-7; PMID:12193640; http://dx.doi.org/ 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- 59. Debeauchamp JL, Moses A, Noffsinger VJ, Ulrich DL, Job G, Kosinski AM, Partridge JF. Chp1-Tas3 interaction is required to recruit RITS to fission yeast centromeres and for maintenance of centromeric heterochromatin. Mol Cell Biol 2008; 28:2154-66; PMID:18212052; http://dx.doi.org/ 10.1128/MCB.01637-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lan F, Zaratiegui M, Villen J, Vaughn MW, Verdel A, Huarte M, Shi Y, Gygi SP, Moazed D, Martienssen RA. S. pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription. Mol Cell 2007; 26:89-101; PMID:17434129; http://dx.doi.org/ 10.1016/j.molcel.2007.02.023 [DOI] [PubMed] [Google Scholar]
- 61. Scott KC, White CV, Willard HF. An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1. PLoS One 2007; 2:e1099; PMID:17971862; http://dx.doi.org/ 10.1371/journal.pone.0001099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res 2002; 62:7213-8; PMID:12499261 [PubMed] [Google Scholar]
- 63. Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 2004; 4:143-53; PMID:14732866; http://dx.doi.org/ 10.1038/nrc1279 [DOI] [PubMed] [Google Scholar]
- 64. Manuyakorn A, Paulus R, Farrell J, Dawson NA, Tze S, Cheung-Lau G, Hines OJ, Reber H, Seligson DB, Horvath S, et al. . Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. J Clin Oncol 2010; 28:1358-65; PMID:20142597; http://dx.doi.org/ 10.1200/JCO.2009.24.5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet 2007; 8:35-46; PMID:17173056; http://dx.doi.org/ 10.1038/nrg2008 [DOI] [PubMed] [Google Scholar]
- 66. Takahashi K, Takayama Y, Masuda F, Kobayashi Y, Saitoh S. Two distinct pathways responsible for the loading of CENP-A to centromeres in the fission yeast cell cycle. Philos Trans R Soc Lond B Biol Sci 2005; 360:595-606; PMID:15897182; http://dx.doi.org/ 10.1098/rstb.2004.1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Uhlmann F. Chromosome cohesion and segregation in mitosis and meiosis. Curr Opin Cell Biol 2001; 13:754-61; PMID:11698193; http://dx.doi.org/ 10.1016/S0955-0674(00)00279-9 [DOI] [PubMed] [Google Scholar]
- 68. Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 2006; 127:983-97; PMID:17129783; http://dx.doi.org/ 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- 69. Niwa O, Matsumoto T, Chikashige Y, Yanagida M. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J 1989; 8:3045-52; PMID:2583093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci 1996; 109:2637-48; PMID:8937982 [DOI] [PubMed] [Google Scholar]
- 71. Cha HJ, Byrom M, Mead PE, Ellington AD, Wallingford JB, Marcotte EM. Evolutionarily repurposed networks reveal the well-known antifungal drug thiabendazole to be a novel vascular disrupting agent. PLoS Biol 2012; 10:e1001379; PMID:22927795; http://dx.doi.org/ 10.1371/journal.pbio.1001379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shih C, Teicher BA. Cryptophycins: a novel class of potent antimitotic antitumor depsipeptides. Curr Pharm Des 2001; 7:1259-76; PMID:11472266; http://dx.doi.org/ 10.2174/1381612013397474 [DOI] [PubMed] [Google Scholar]
- 73. Burris HA, 3rd. Docetaxel in combination with fluorouracil for advanced solid tumors. Oncology (Williston Park) 1997; 11:50-2; PMID:9364544 [PubMed] [Google Scholar]
- 74. Zhou Y, Lin J, Johnson RG., Jr KOS-862 (epothilone D) and 5′-deoxy-5-fluorouridine or 5-fluorouracil in human breast cancer cells: marked synergistic antitumor effects and induction of thymidine phosphorylase expression. Proc Am Assoc Cancer Res 2004; 45:Abstr 5429. [Google Scholar]
- 75. Kniola B, O’Toole E, McIntosh JR, Mellone B, Allshire R, Mengarelli S, Hultenby K, Ekwall K. The domain structure of centromeres is conserved from fission yeast to humans. Mol Biol Cell 2001; 12:2767-75; PMID:11553715; http://dx.doi.org/ 10.1091/mbc.12.9.2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bernstein E, Allis CD. RNA meets chromatin. Genes Dev 2005; 19:1635-55; PMID:16024654; http://dx.doi.org/ 10.1101/gad.1324305 [DOI] [PubMed] [Google Scholar]
- 77. Cam HP, Chen ES, Grewal SI. Transcriptional scaffolds for heterochromatin assembly. Cell 2009; 136:610-4; PMID:19239883; http://dx.doi.org/ 10.1016/j.cell.2009.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol 2010; 17:251-7; PMID:20118936; http://dx.doi.org/ 10.1038/nsmb.1741 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.