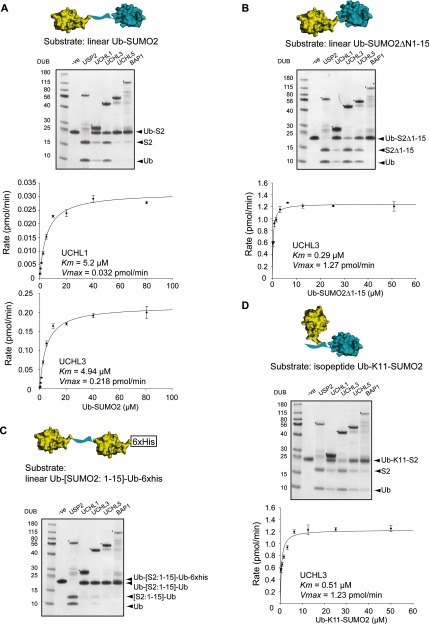
Figure 5. Ub-SUMO2 substrate requirements for cleavage with UCH enzymes.
(A) Linear Ub-SUMO2 substrate (structural representation above) was incubated with UCH DUBs and the reaction products were fractionated by SDS/PAGE and visualized by Coomassie Blue staining (top panel). Michaelis–Menten kinetic analysis of UCH-L1 and UCH-L3 with Ub-SUMO2 as substrate (bottom panels). (B) Structural representation of Ub-SUMO2ΔN1-15 used as substrate. DUB activity was assessed as in (A). Michaelis–Menten kinetic analysis of UCH-L3 with Ub-SUMO2ΔN1-15 as substrate. (C) Structural representation of Ub-[SUMO2 1:15]-Ub-6xHis used as substrate. DUB activity was assessed by release of free Ub and free [SUMO2:1-15]-Ub or cleavage of the 6-His tag. (D) Structural representation of isopeptide-linked Ub-K11-SUMO2 used as substrate. DUB activity was assessed as in (A). Michaelis–Menten kinetic analysis of UCH-L3 with Ub-K11-SUMO2 as substrate. Asterisks (*) denote the DUB in each reaction. Structural representation of each Ub-SUMO2 substrate was created by using the Ub structure (PDB: 1UBQ, yellow) [46] and the SUMO2 structure (PDB: 1WM2, blue) [47] (the flexible N-terminus of SUMO2 is represented by a blue ribbon).