Table 3.
Enzyme substrate promiscuity indices
| Enzymeb | Standard Index of substrate promiscuity (I)ac | Weighted Index of substrate promiscuity (J)ac | ||||||
|---|---|---|---|---|---|---|---|---|

|
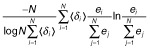
|
|||||||
| I | I-ValP | I-Pyr | I-P5C | J | J-ValP | J-Pyr | J-P5C | |
| Cgl | 0.71 | 0.57 | 0.74 | 0.75 | 0.61 | 0.39 | 0.61 | 0.69 |
| Spr | 0.67 | 0.71 | 0.70 | 0.70 | 0.59 | 0.67 | 0.60 | 0.64 |
| Svi1 | 0.67 | 0.62 | 0.70 | 0.70 | 0.59 | 0.59 | 0.60 | 0.63 |
| Svi2 | 0.47 | 0.81 | 0.49 | 0.49 | 0.42 | 0.79 | 0.43 | 0.46 |
| Sgr | 0.65 | 0.66 | 0.68 | 0.67 | 0.57 | 0.64 | 0.58 | 0.61 |
| Sav | 0.70 | 0.77 | 0.73 | 0.73 | 0.62 | 0.74 | 0.64 | 0.67 |
| Sam | 0.74 | 0.66 | 0.77 | 0.77 | 0.66 | 0.64 | 0.66 | 0.70 |
| Sco2 | 0.83 | 0.75 | 0.83 | 0.83 | 0.77 | 0.73 | 0.76 | 0.83 |
| Sli2 | 0.80 | 0.71 | 0.83 | 0.83 | 0.75 | 0.70 | 0.75 | 0.78 |
| Sco1 | 0.64 | 0.42 | 0.67 | 0.67 | 0.56 | 0.40 | 0.57 | 0.61 |
| Sli1 | 0.64 | 0.51 | 0.67 | 0.67 | 0.56 | 0.48 | 0.56 | 0.60 |
| Sco_P5CR | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Both indices range between 0 and 1.
Enzyme nomenclature is as in Table 1.
N, number of substrates; e, catalytic efficiency; δi = δij / δset, δij, the mean Tanimoto distance from a member i to all the other members in the set; δset, the overall set dissimilarity.
