Figure 1.
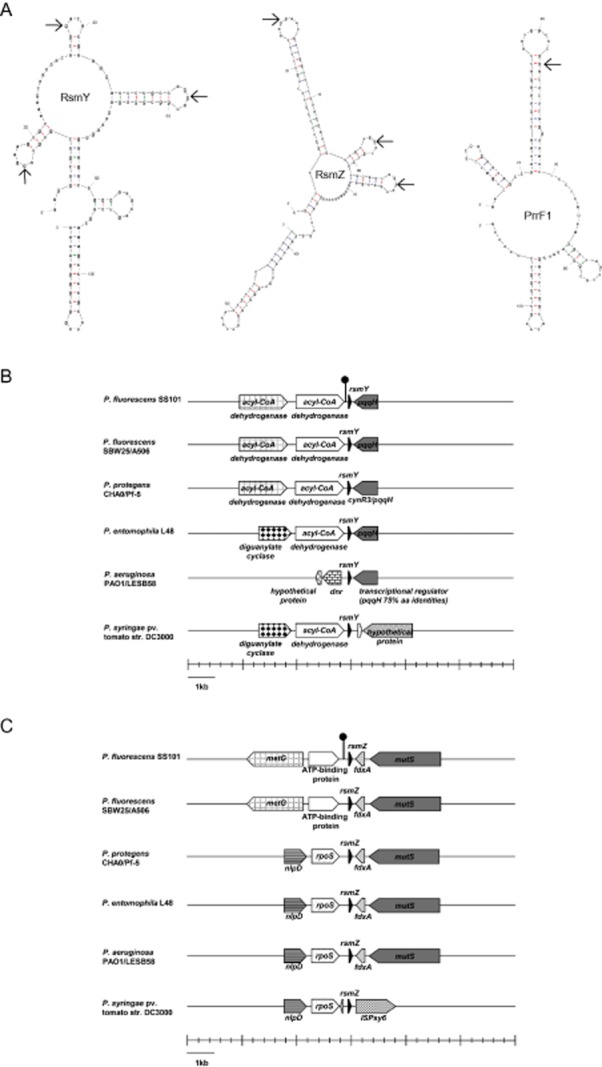
Secondary structures of small RNAs, RsmY, RsmZ, PrrF1 in P. fluorescens SS101 and the genetic organization of rsmY and rsmZ in strain SS101 and other Pseudomonas species and strains.A. Predicted secondary structures of RsmY, RsmZ and PrrF1 of P. fluorescens SS101 by MFOLD (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form). The typical GGA motifs located in the loop regions are indicated with arrows.B. Genetic organization of rsmY regions in different Pseudomonas species and strains. Block arrows indicate directionality of the open reading frame, and orthologous genes are represented by color and pattern. The loop symbol in front of rsmY indicates the position of the upstream activating sequence (UAS for rsmY: TGTAAGCATTCTCTTACA). Abbreviations: pqqH/cynR3/dnr: transcriptional regulator.C. Genetic organization of rsmZ regions in different Pseudomonas species and strains. Block arrows indicate directionality of the open reading frame, and orthologous genes are represented by colour and pattern. The loop symbol in front of rsmZ indicates the position of the UAS (UAS for rsmZ: TGTAAGCATTCGCTTACT). Abbreviations: metG: methionyl-tRNA synthetase; fdxA: ferredoxin; mutS: DNA mismatch repair protein; nlpD: lipoprotein; rpoS: RNA polymerase sigma factor; ISPsy6: transposase.
