Figure 1.
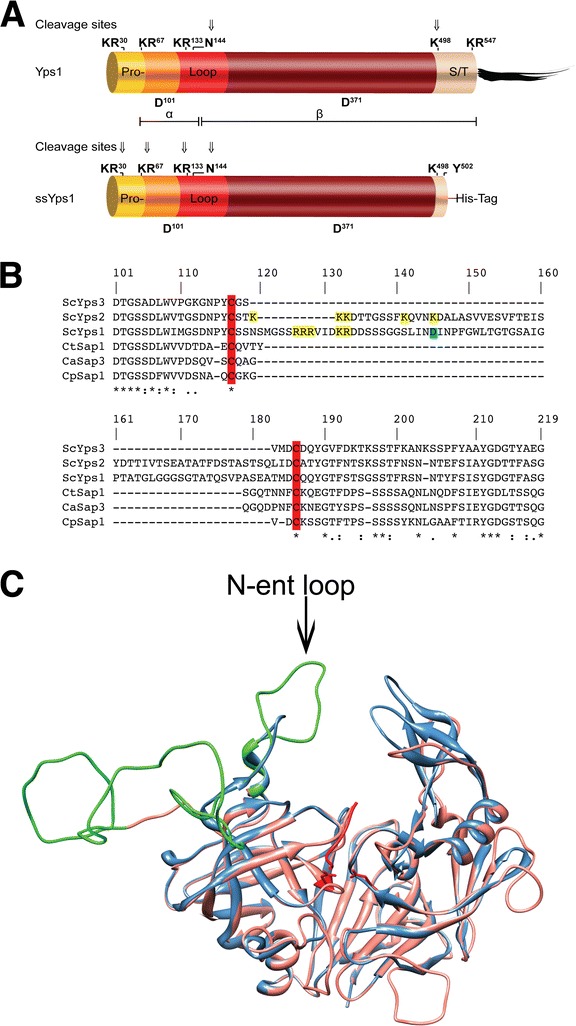
Alignment and structure of ScYps1 vs CaSap3. A) Schematic representation of native Yps1 and ssYps1 (adapted from Gagnon-Arsenault et al., [8]). Cleavage sites known to be used in each forms are indicated by arrows. B) ClustalW alignment [20] of S. cerevisiae yapsins Yps3 (ScYps3; NP_013222.1), Yps2 (ScYps2; NP_010428.3) and Yps1 (ScYps1; NP_013221.1) with Secreted Aspartyl Proteinases Sapt1 from C. tropicalis (CtSap1; Q00663.1), Sap3 from C. albicans (CaSap3; XP_723210.1) and Sapp1 from C. parapsilosis (CpSap1; ACL81524.1) displaying the insertion loop located between the conserved cysteine residues (highlighted in red). The basic residues (highlighted in yellow) present in the insertion loop of ScYps1 and ScYps2 are shown along with the known N-terminus of the fastest migrating form of Yps1 β subunit (highlighted in blue). C) Modelled chimeric structure of ScYps1 (residues 76 to 490, in blue) and CaSap3 (residues 66 to 398, in orange) obtained from the ModWeb server. The N-ent loop (indicated by the arrow) of Yps1 is shown in green. The aspartic residues of the catalytic center and the inhibitor Pepstatin A are shown in red.
