Abstract
Background
Although single photon emission computed tomography myocardial perfusion imaging (SPECT MPI) has improved the diagnosis and risk stratification of patients with suspected coronary artery disease, it remains a primary source of low dose radiation exposure for cardiac patients. To determine the biological effects of low dose radiation from SPECT MPI, we measured the activation of the DNA damage response pathways using quantitative flow cytometry and single cell gene expression profiling.
Methods and Results
Blood samples were collected from patients before and after SPECT MPI (n=63). Overall, analysis of all recruited patients showed no marked differences in the phosphorylation of proteins (H2AX, p53, and ATM) following SPECT. The majority of patients also had either down-regulated or unchanged expression in DNA damage response genes at both 24 and 48 hours post-SPECT. Interestingly, a small subset of patients with increased phosphorylation also had significant up-regulation of genes associated with DNA damage, whereas those with no changes in phosphorylation had significant down-regulation or no difference, suggesting that some patients may potentially be more sensitive to low dose radiation exposure.
Conclusions
Our findings showed that SPECT MPI resulted in a variable activation of the DNA damage response pathways. Although only a small subset of patients had increased protein phosphorylation and elevated gene expression post-imaging, continued care should be taken to reduce radiation exposure to both patients and operators.
Keywords: radiation risk, SPECT, DNA damage, protein phosphorylation, gene expression
The utilization of radiation-producing imaging tests has risen dramatically in recent years, especially those used to risk stratify and manage patients with coronary artery disease (CAD).1 Single photon emission computed tomography myocardial perfusion imaging (SPECT MPI) test is the most common diagnostic imaging procedure performed in patients with CAD, and its usage has doubled in the last decade alone.2 These staggering numbers have raised concern about the biological effects of low dose radiation exposure for patients, physicians, and technical staff members. However, determining the radiation risk after low dose radiation exposure (<100 mGy) remains challenging.3 Since the 1970s, the International Commission on Radiological Protection (ICRP) has recommended extrapolating the linear no-threshold (LNT) model, which is based on epidemiological studies in those exposed to high dose radiation, to estimate low dose radiation risk. The LNT model states that radiation risk is directly proportional to dose; thus, any small amount of radiation is potentially harmful.4-6
Recent studies, however, have questioned the validity of the LNT model for evaluating radiation risk at low doses.3, 7-9 Measuring the cellular effects of radiation has emerged as an alternative strategy of assessing radiation risk. Exposure to high dose ionizing radiation results in the development of double stranded breaks (DSBs) in cells. In response to DNA damage, cells activate proteins and genes involved in apoptosis, DNA repair, cell cycle regulation, and chromatin remodeling, a process collectively known as the DNA damage response pathways.10, 11 The extent to which these pathways are induced or repressed influences how patients respond to radiation exposure. Whether low dose radiation exposure from SPECT MPI activates these pathways has not been fully explored. In the present study, we examined the effects of low dose radiation on the DNA damage response pathways in T-lymphocytes isolated from adult patients undergoing SPECT MPI using quantitative flow cytometry and single cell gene expression profiling.
Methods
Patient Population and Clinical Imaging Studies
For this study, we recruited a total of 90 adult patients aged ≥18 years, who underwent a clinically indicated SPECT imaging examination (n=63), invasive coronary angiogram (n=12; as a positive control), or echocardiography (n=15; as a negative control) at the Veterans Affairs Palo Alto Health Care System (Figure 1). For SPECT MPI, patients underwent a Technetium-99m (Tc-99m) tetrofosmin rest/exercise same day protocol (Figure 2A). The mean injected dose during rest was 6.9±0.5 millicuries (mCi). For the stress study, the dose averaged 23.8±1.4 mCi. The injection of a second dose of radionuclide occurred within ~1.5 hours of the first dose. All examinations were performed using an Infinia Hawkeye 3.0 (GE, Milwaukee, WI). Coronary catheterization was performed in standard views using the Innova Interventional X-ray system (GE). All transthoracic echocardiograms were performed using a VIVID7 ultrasound machine (GE). Demographic and clinical information was obtained from the electronic medical record. Informed consent was obtained from all patients.
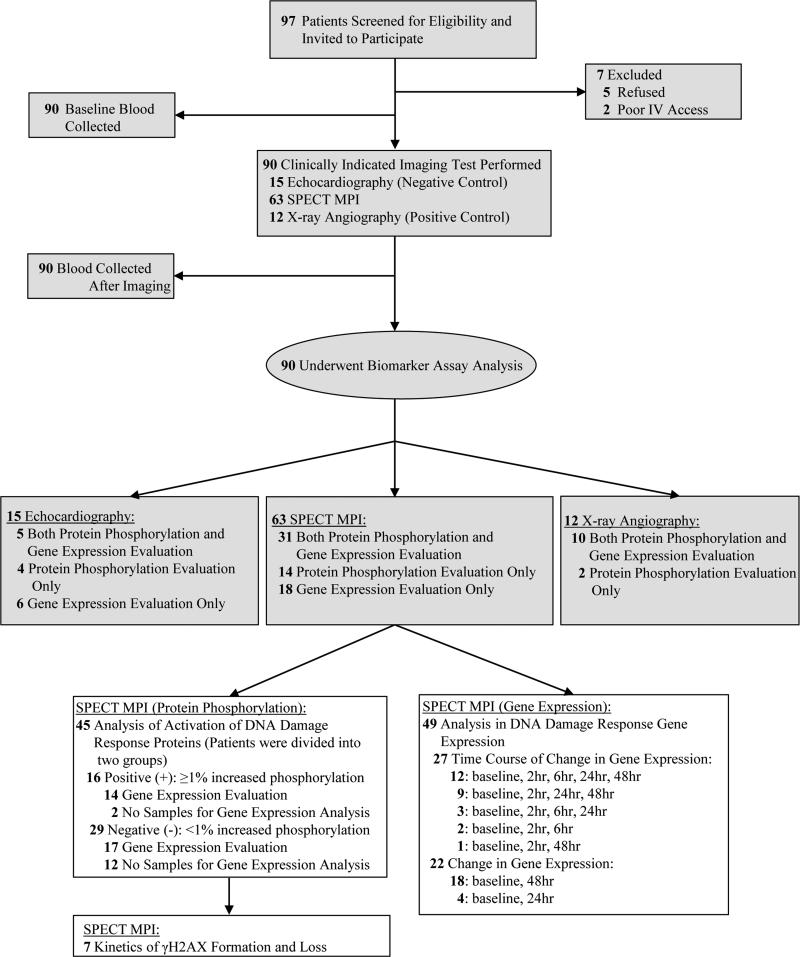
Figure 1.
Flow chart of the study showing the number of patients enrolled in the study. SPECT: single photon emission computed tomography; MPI: myocardial perfusion imaging; IV: intravenous.
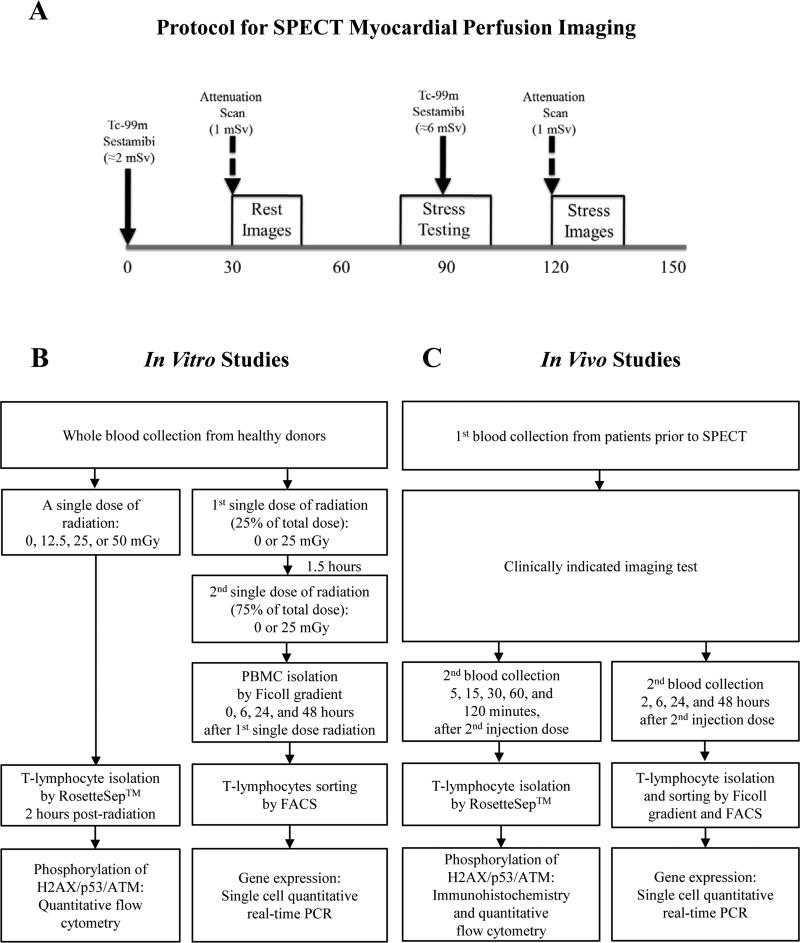
Figure 2.
Schematic diagram of (A) SPECT MPI protocol and overall (B) in vitro and (C) in vivo study design. PBMC: peripheral blood mononuclear cell; FACS: fluorescence-activated cell sorting.
Sample Collection
For the in vitro study, peripheral blood from healthy volunteers was drawn into vacutainer tubes containing EDTA and placed immediately on ice prior to irradiation. Sixteen cc of whole blood from healthy volunteers (4 cc per each irradiation dose, n=13) was collected for evaluation of proteomic changes (Figure 2B). To determine gene expression, 12 cc of blood (3 cc at each time point) was collected from individual donors (n=3).
For the in vivo study, blood samples were collected from each patient prior to and after the medical imaging procedure (Figure 2C). DNA repair was then arrested by placing the samples immediately on ice. Specifically, for SPECT MPI, we obtained blood samples at 2 hours after the first injection dose (i.e., approximately 30 minutes after the second injection dose), and the percent of cells expressing phosphorylated DNA damage markers was measured by quantitative flow cytometry. In a subset of patients, additional samples were taken after the second injection dose to examine the kinetics of γH2AX formation, and loss over time by quantitative flow cytometry (e.g., at 15 and 30 minutes) and immunocytochemistry (e.g., at 5, 15, 30, 60, and 120 minutes). To determine the changes in gene expression after SPECT MPI, blood samples were obtained at baseline and then at 2, 6, 24, and 48 hours after the second injection dose. Expression of selected DNA damage response genes was assessed by quantitative single cell real-time PCR. In addition, blood samples were collected from patients at similar time points before and after invasive X-ray angiography or echocardiography for protein phosphorylation and gene expression analysis, respectively.
Estimation of Radiation Dose
Effective dose equivalent from SPECT was calculated from the injected radiotracer dose according to the ICRP Publication 106, using the following conversion factors: 1) Rest = 0.0080 mSv/MBq and 2) Stress = 0.0069 mSv/MBq.12 Effective dose equivalent from invasive X-ray angiography was calculated according to a previous report (0.22 mSv/Gy·cm2).13 Total dose area product (DAP), measured in cGy·cm2, is included in Supplemental Table 1.
Quantitative Flow Cytometry
T-lymphocytes were isolated from whole blood using the RosetteSep™ Human T Cell Enrichment Cocktail (StemCell Technologies, Inc., Vancouver, Canada), according to the manufacturer's protocol. Briefly, blood samples were incubated with RosetteSep™ Human T Cell Enrichment Cocktail for 20 minutes at room temperature. After density gradient centrifugation on Ficoll-Paque™ Plus (Amersham/GE Healthcare, Piscataway Township, NJ), the enriched T-lymphocytes expressing CD3+ were collected from the interface between the density medium and plasma. After fixation, cell permeabilization was carried out on ice for 10 minutes. Total and phosphorylated forms of H2AX were determined by labeling cells with either anti-histone H2AX-FITC or anti-phospho histone H2AX-PerCP (FlowCellect™ Dual Detection kits, Millipore, Billerica, MA) for 30 minutes in the dark at room temperature. To measure the phosphorylated form of ataxia telangiectasia mutated (ATM) and tumor protein 53 (p53), cells were labeled with anti-phospho ATM-PE (Millipore, Billerica, MA) or anti-phospho p53-FITC (Cell Signaling Technologies, Inc., Danvers, MA) in the dark on ice for 60 minutes, respectively. Finally, cells were resuspended in fluorescence activated cell sorting (FACS) buffer (DPBS with 2% FBS and 2 mM EDTA) and analyzed using a LSRII flow cytometer (BD Biosciences, San Jose, CA). Fluorochrome and isotype matched controls (PerCP-IgG1, FITC-IgG, and PE-IgG1κ; Millipore, Billerica, MA) as well as unlabeled samples were used to set the appropriate gate parameters and served as negative controls (data not shown). A total of 10,000 events were recorded in each analysis. Data analysis was conducted using the FlowJo software (Tree Star, Ashland, OR).
Single Cell Quantitative Real-Time PCR
Peripheral blood mononuclear cells (PBMCs) were isolated from the buffy coat after density gradient centrifugation on 1.077g/ml Ficoll-Paque™ Plus. Freshly isolated PBMCs were pre-incubated with 20 μl of FcR-blocking reagent (Miltenyi, Auburn, CA) and stained with 5 μl of Alexa Fluor® 647 mouse anti-human CD3 antibody (BD Biosciences) by incubating cells in 100 μl of FACS buffer on ice for 20 minutes. The specificity of staining was confirmed by using an isotype-matched control antibody (Alexa Fluor® 647-IgG1κ; BD Biosciences). Dead cells were excluded by adding 7-Amino-Actinomycin D (7-AAD) to the cell suspension prior to sorting. Using a FACS Aria (BD Biosciences), cells (CD3+ and 7AAD−) were sorted directly into 96-well 0.2 ml PCR plates, containing buffers and enzymes for reverse transcription according to the manufacturer's instructions (Fluidigm, South San Francisco, CA), as described previously.14 TaqMan® Gene Expression Assay Reagents for DNA damage response genes and 18S (housekeeping gene) were used as specific probes and primers for PCR amplifications (Supplemental Table 2). Reverse transcription and specific amplification of individual genes defining DNA damage responses were performed using the following protocol: 50°C for 15 minutes, 70°C for 2 minutes, 95°C for 15 seconds, and 60°C for 4 minutes, repeated 18 times. Quantitative real-time PCR was then conducted on a Fluidigm 48×48 Dynamic Array microfluidic chip by partitioning the samples into 48 microfluidic chambers, running in a BioMark HD reader, and using the Fluidigm Real-time PCR analysis software. Results are shown as threshold cycles (CT), which measures target transcript abundance in the samples and is analyzed using an equation that was described previously.15 All reactions were performed in duplicates or triplicates.
Immunocytochemistry
Isolated T-lymphocytes (~1.6×105 cells/150 μl per slide) were spun onto Superfrost/Plus microscope slides (Fisher Scientific, Pittsburgh, PA) using the Cytospin 4 (Thermo Scientific; Waltham, MA) at 500 rpm for 5 minutes at low acceleration. Cells were fixed in 4% paraformaldehyde for 15 minutes at room temperature. After rinsing with PBS, cells were blocked, and permeabilized in 5% bovine serum albumin (BSA) and 0.2% Triton X-100 for an hour at room temperature, followed by incubation with primary antibodies, including mouse anti-γH2AX monoclonal antibody (Millipore, Billerica, MA) and rat anti-CD3 monoclonal antibody (AbDSerotec, Düsseldorf, Germany) diluted in antibody diluent buffer (IHC World, Woodstock, MD) overnight at 4°C. Cells were then washed with 0.2% Tween-20 in PBS. After additional rinsing with PBS, cells were incubated in the dark for an hour with a secondary antibody, either goat-anti-mouse IgG conjugated with Alexa Fluor 488 or goat-anti-rat IgG conjugated with Alexa Fluor 594. The slides were washed and nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI). The number of γH2AX (phosphorylated at serine 139) foci per lymphocyte was determined using a fluorescence microscope with 40 x magnification. Forty cells were counted by two blinded observers.
Statistical Analysis
All statistical analysis of data was completed using SigmaStat 3.5 (SPSS Inc., Chicago, IL). Changes in protein phosphorylation and gene expression before and after medical imaging (i.e., SPECT MPI, invasive X-ray angiography, and echocardiography) were compared with a paired t-test and Wilcoxon signed-rank test with Bonferroni correction for normally and non-normally distributed data, respectively. To test for serial changes in protein activation and gene expression, a one-way repeated-measures (RM) ANOVA with Bonferroni post-hoc analysis was used. If data were not normally distributed, the Friedman RM ANOVA, followed by Wilcoxon signed-rank test with Bonferroni correction, was conducted.
For analysis of individual variation in activation of DNA damage response proteins, patients were divided into two groups: 1) those with marked increased phosphorylation (+) and 2) those with no marked change (−). A marked increase in phosphorylation was defined as an increase in phosphorylation of at least 1% (≥100 events) compared to baseline. By contrast, the negative control group showed a minor increase of <1% positive cells before and after echocardiography. For analysis of individual variation in gene expression, patients were grouped into the following categories: 1) those with marked up-regulation (Up), 2) those with marked down-regulation (Down), and 3) no marked change (No change). Marked up-regulation and down-regulation were defined as ≥1.5-fold increase and ≥1.5-fold decrease (i.e., ≤0.67 change) in gene expression, respectively.
All values were expressed as mean ± SEM and statistical probability of adjusted P<0.05 (Bonferroni correction) was considered significant. Bonferroni correction was used for multiple comparisons, with P<0.05 (number of comparisons) being considered significant.
Results
Demographics and Clinical Characteristics
A total of 63 patients were recruited for the evaluation of protein phosphorylation (n=45) and/or gene expression (n=49) after SPECT MPI (Figure 1). Thirty-one patients underwent evaluation for both changes in protein phosphorylation and gene expression, whereas 14 and 18 patients were evaluated for either protein phosphorylation or gene expression, respectively. Serial blood samples were collected in a subset of 7 patients to evaluate the kinetics of γH2AX formation and loss using immunohistochemistry. The time course of gene expression was also analyzed by collecting serial blood samples from a subset of 27 patients (baseline, 2, 6, 24, and 48 hours post-SPECT). The majority of patients undergoing SPECT MPI were Caucasian (73.0%), male (95.2%), and overweight (average BMI: 30.7±9.6 kg/m2) with significant previous or active smoking history (74.6%), but no history of prior malignancy (66.7%). The average age was 69.6±10.5 years. Most patients underwent exercise testing (55.6%), with the remainder undergoing pharmacological stress testing. Average rest, stress, and total effective doses were 2.7±0.6 mSv, 8.1±0.4 mSv, and 10.7±0.4 mSv, respectively. In addition, as a positive control, a total of 12 patients were recruited for the evaluation of protein phosphorylation (n=12) and gene expression (n=10) after invasive X-ray angiography. Samples were also collected from a total of 15 patients undergoing echocardiography (as a negative control) for analysis of protein phosphorylation (n=9) and gene expression (n=11), respectively. The demographics and clinical characteristics for the invasive X-ray angiography and echocardiography patients are included in the Supplemental Table 1 and were not significantly different from the patients undergoing SPECT MPI.
Phosphorylation of DNA Damage Marker Proteins after SPECT MPI
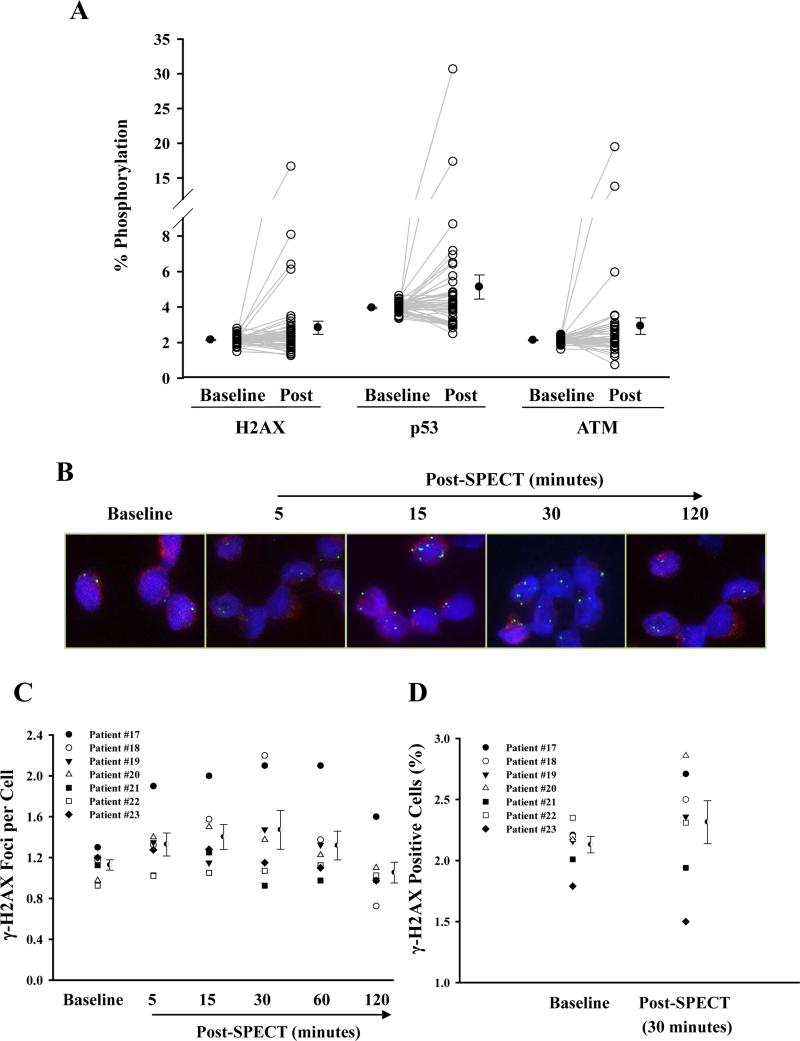
To examine the effects of radiation exposure from SPECT MPI, we measured the phosphorylation of three DNA damage marker proteins (H2AX, p53, and ATM) which are often used as indicators of radiation-induced cellular damage.4, 16-18 We collected T-lymphocytes from patients before and after SPECT. Overall, analysis of all recruited patients (n=45, closed circle) showed no marked differences in phosphorylation for all three proteins following SPECT when compared with baseline value (Figure 3A). Analysis of individual responses, however, showed that some patients (i.e., 35.6%, n=16/45, open circle) had increased protein phosphorylation post-SPECT (Figure 3A). Consistent with these findings, blood samples drawn from healthy volunteers (n=4-8) subjected to a single dose of in vitro radiation (of varying intensity) showed no marked changes in phosphorylation of measured proteins at lower doses (12.5 and 25 mGy, closed squares). This is equivalent to the amount of radiation exposure from SPECT MPI, which is approximately ~10 mGy (Supplemental Figure 1). Variation in individual response to low dose radiation, however, was also observed (open circles).
Figure 3.
The phosphorylation of DNA damage-marker proteins in T-lymphocytes isolated from patients undergoing SPECT MPI. T-lymphocytes were isolated from patients before (baseline) and after SPECT MPI (post). (A) The percentage of phosphorylated cells for H2AX, p53, and ATM in T-lymphocytes taken from patients undergoing SPECT MPI (n=45). Each individual data point (open circle) represents a separate patient sample from 45 pateints, and closed circles represent mean ± SEM taken at baseline and post-SPECT MPI. Total and phosphorylated H2AX, p53, and ATM in T-lymphocytes were measured by flow cytometry. (B) Representative fluorescence microscopy images of γ-H2AX foci (green) and (C) the mean number of foci per cell isolated in T cell lymphocytes at baseline, 5, 15, 30, 60, and 120 minutes following SPECT measured by immonohistochemistry. Semicircle represents mean ± SEM for each group (n=7). (D) Flow cytometric analysis of γ-H2AX positive T-lymphocytes before and 30 minutes after SPECT. Semicircle represents mean ± SEM for each group (n=7).
To evaluate the sensitivity of quantitative flow cytometry for detecting small changes in phosphorylation in vivo and to verify that peak changes in phosphorylation occurred at 30 minutes in most patients, we next measured serial changes in protein phosphorylation using immunohistochemistry (n=7), a method that has been shown to detect these changes after exposure to radiation doses as low as 1.2 mGy in vitro.19 Consistent with other studies,4, 20 γ-H2AX foci appeared as early as 5 minutes post-radiation, reached a maximum at 15-30 minutes, and decreased thereafter (Figure 3B-C). No significant differences were found in the change in phosphorylation of H2AX by immunohistochemistry (Figure 3C) and quantitative flow cytometry (Figure 3D) compared to baseline, defined as an excess of 1 foci per cell and an 1% increase in events, respectively, although considerable variation was seen among patients using both assays (Supplemental Table 3). Importantly, the two observers showed a moderately high agreement for counting the average foci per cell for each immunohistochemistry sample (Intraclass correlation coefficient = 0.763).
Interestingly, T-lymphocytes isolated from patients after invasive X-ray angiography, an imaging modality that exposes patients to short bursts of radiation that were on average higher than SPECT MPI (average dose: 18.2±10.6 mSv, Supplemental Table 1), showed significantly higher phosphorylation of H2AX (3.61±0.50% vs. 2.14±0.04%, P=0.004), p53 (6.68±1.42% vs. 3.75±0.10%, P=0.015), and ATM (7.44±1.99% vs. 2.05±0.05%, P=0.004) compared with baseline (Supplemental Figure 2A). Moreover, all patients undergoing invasive X-ray angiography had activation of at least one marker of DNA damage (Supplemental Figure 2C). By contrast, we found no significant changes in the percent phosphorylation of DNA damage-marker proteins in T-lymphocytes isolated from patients undergoing echocardiography compared to baseline (Supplemental Figure 2B and 2D), supporting the hypothesis that only radiation-producing imaging tests activate the DNA damage response pathways.
Changes in mRNA Expression of DNA Damage Response Genes after SPECT MPI
Because exposure to radiation is known to modulate gene expression, we selected 21 candidate genes based on previous microarray studies performed on human lymphocytes in response to a range of low dose irradiation (0-100 mGy).21-24 In vitro single cell quantitative real-time PCR assays were performed to investigate the temporal expression of these genes in human T-lymphocytes following exposure to fractionated doses simulating SPECT imaging (e.g., 25 mGy). We found an insignificant trend toward down-regulation of 9 DNA damage response genes compared with sham-irradiated control (Supplemental Figure 3), with peak changes mainly occurring 24 hours post-exposure before returning to basal levels at 48 hours. Importantly, there was no difference in cell viability after low radiation exposure, suggesting that T-lymphocytes had a lower expression of these genes rather than a lower number of cells expressing these genes (data not shown).
Subsequently, blood samples were collected from patients before (baseline, n=49), 2 hours (2.1±0.3 hours, n=27), 6 hours (6.5±1.4 hours, n=17), 24 hours (21.8±6.7 hours, n=28), and 48 hours (47.0±0.4 hours, n=40) post-SPECT, and expression of selected genes based on our in vitro experiments were determined. Serial blood samples were initially collected to determine the optimal timing for gene expression analysis. Changes in gene expression were detectable as early as 2 hours post-radiation and in some patients were extended to 48 hours (Supplemental Figure 4). In addition, most patients showed a consistent pattern of up-regulation, down-regulation, or no regulation of each gene, with most changes still detectable up to 24 and/or 48 hours (Supplemental Table 4), forming the basis for subsequent sample collection from remaining patients up to those time points only. No consistent patterns of expression were found across genes in the same patient.
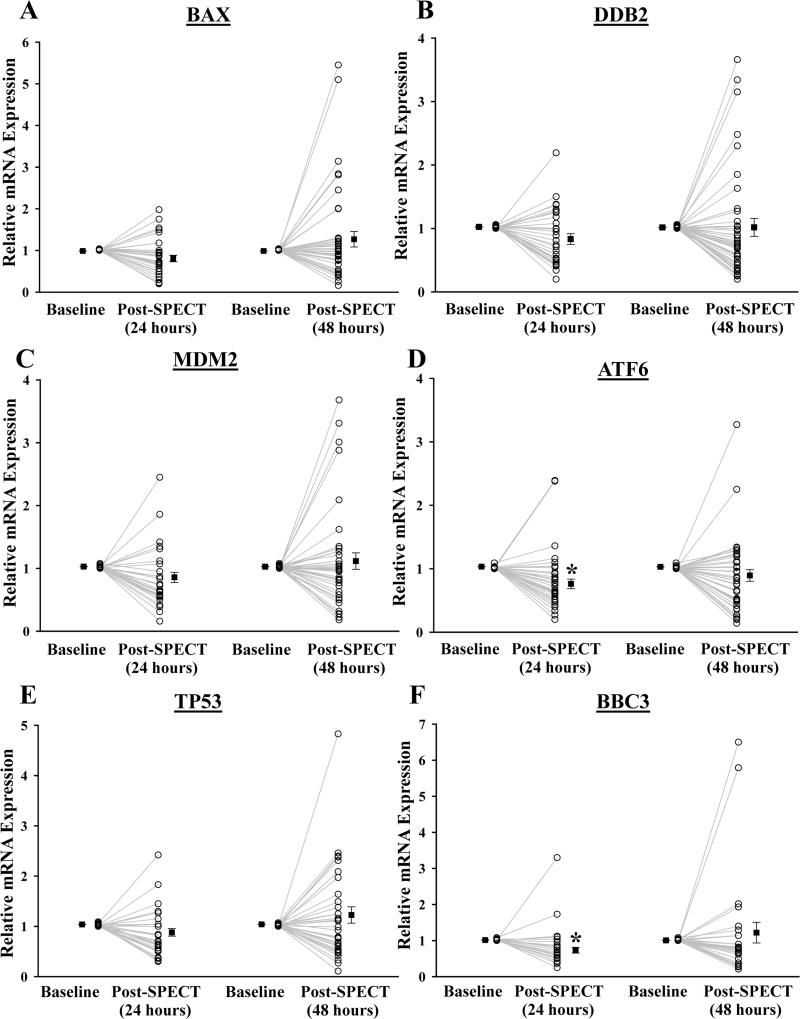
Analysis of the entire cohort (n=49) revealed that mRNA expression varied significantly among individuals (Figure 4), similar to changes in phosphorylation. On average, single cell gene expression analysis of T-lymphocytes at 24 hours post-SPECT demonstrated a trend toward down-regulation of selected genes (Bax: 0.85-fold; Ddb2: 0.84-fold; Mdm2: 0.83-fold; and Tp53: 0.86-fold), although this was not statistically significant except for Atf6 (0.74-fold, P=0.002) and Bbc3 (0.66-fold, P=0.012). Changes in gene expression were still detectable at 48 hours in some patients, but appeared to return to basal expression in the majority of patients, in line with our in vitro findings. To further assess overall patterns of gene changes over time in patients after SPECT, patients were split into three groups according to the relative mRNA expression in each gene, classified as Down (≥1.5-fold decrease), Up (≥1.5-fold increase), or No change (Supplemental Figure 5). The majority of patients (~70-95%) had either down-regulated or unchanged expression in DNA damage response genes at both 24 and 48 hours post-SPECT.
Figure 4.
The relative mRNA expression of DNA damage response genes in T-lymphocytes isolated from patients undergoing SPECT MPI. T-lymphocytes were isolated from patients before (baseline, n=49), 24 hours (n=28), and 48 hours (n=40) after SPECT MPI. Relative mRNA expression levels of (A) Bax, (B) Ddb2, (C) Mdm2, (D) Atf6, (E) Tp53, and (F) Bbc3 in T-lymphocytes were determined by single cell quantitative real-time PCR. Each individual data point represents a separate patient sample taken from baseline and after SPECT MPI (open circle). Closed squares shown are displayed as mean ± SEM for each group. *Statistically significant from baseline (Bonferroni-adjusted P<0.05).
Of note, invasive X-ray angiography resulted in a significantly elevated expression of most genes (Bax: 2.05-fold, P=0.048; Ddb2: 2.19-fold, P=0.036; Mdm2: 1.83-fold, P=0.036; Tp53: 2.35-fold, P=0.048; and Bbc3: 2.46-fold, P=0.024) in T-lymphocytes collected from patients at 24 hours (Supplemental Figure 2E). Results from negative control samples taken from patients at 24 and 48 hours following echocardiography showed no significant changes in gene expression (Supplemental Figure 2F).
Correlation between Protein Phosphorylation and mRNA Expression of DNA Damage Response Genes
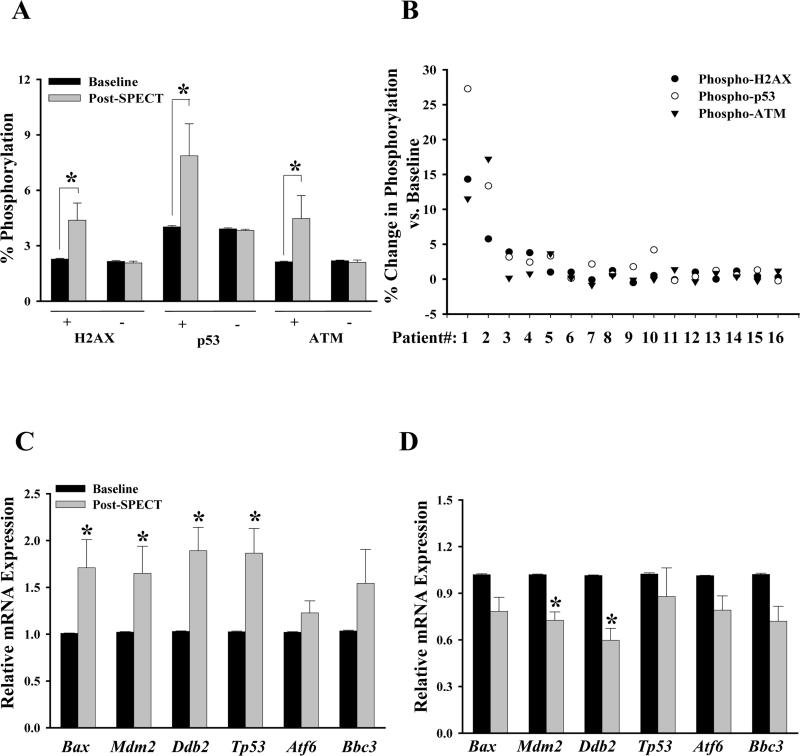
Considering that a small fraction of patients showed increased protein phosphorylation and elevated gene expression of DNA damage response genes in response to SPECT-MPI, we determined the correlation between protein activation and mRNA expression of DNA damage response genes. We first separated patients (n=45, Figure 3A) into two groups based on their phosphorylation positivity (“+” represents phosphorylation ≥1%, whereas “–” represents phosphorylation <1%). Sixteen out of 45 patients (“group +”) had marked increases in protein phosphorylation (γ-H2AX: 4.40±0.93% vs. 2.30±0.04%, P=0.006; phospho-p53: 7.90±1.72% vs. 4.00±0.07%, P=0.002; phospho-ATM: 4.47±1.24% vs. 2.13±0.04%, P=0.012; n=16, Figure 5A), whereas the remaining patients (“group –”) had little or no change (n=29, Figure 5A). These results suggest that some patients may be more susceptible to cellular damage following low dose radiation exposure. Of these 16 positive patients, only 3 showed an increase in all 3 proteins compared to baseline (Figure 5B). Both subgroups of patients did not differ with respect to demographic or clinical characteristics (Table).
Figure 5.
Correlation between protein phosphorylation and mRNA expression of DNA damage response genes. (A) Bar graphs showing the percentage of phosphorylated cells for H2AX, p53, and ATM among individual patients with ≥1% (+, n=16) and <1% (-, n=29) before and after SPECT MPI. Data shown are mean ± SEM for each group. *Statistically significant from baseline (Bonferroni-adjusted P<0.05). (B) Individual variation in the pattern of protein activation in patients showing positive phosphorylation (n=16). Bar graphs depicting the changes in gene expression in T-lymphocytes isolated from patients with (C) and without (D) evidence of activation of DNA damage response proteins. Data shown are mean ± SEM for each group (C: n=14 and D: n=17). *Statistically significant from baseline (Bonferroni-adjusted P<0.05).
Table.
Demographic and clinical characteristics in patients undergoing SPECT MPI based on changes in phosphorylation.
| ≥ 1% Increase (n=16) | <1% Increase (n=29) | P value | |
|---|---|---|---|
| Age (years) | 68.4 ± 6.6 | 69.2 ± 10.1 | 0.74 |
| Men | 78.6 ± 6.7 | 68.8 ± 9.8 | 0.62 |
| Women | 63.0 ± 00 | 75.0 ± 12.0 | n/a |
| Race (%) | |||
| Caucasian | 75.0 (12/16) | 69.0 (20/29) | 0.74 |
| Clinical Factors | |||
| Average BMI (kg/m2) | 28.7 ± 10.9 | 30.7 ± 12.0 | 0.34 |
| History of Smoking (%) | 68.8 (11/16) | 65.5 (19/29) | 1.00 |
| History of Cancer (%) | 18.8 (3/16) | 20.7 (6/29) | 1.00 |
| Type of Stress (%) | |||
| Exercise | 50 (8/16) | 65.5 (19/29) | 0.35 |
| Pharmacological | 50 (8/16) | 34.5 (10/29) | 0.35 |
| Total Radiation Dose (mSv) (including CT attenuation scan) | 10.0 ± 0.4 | 10.0 ± 0.4 | 0.83 |
| Rest | 3.0 ± 0.2 | 3.0 ± 0.4 | 0.69 |
| Stress | 7.0 ± 0.2 | 7.1 ± 0.4 | 0.51 |
Data presented as mean ± SD or percentages. BMI, body mass index; kg, kilogram; mSv, millisievert.
Interestingly, patients with increased phosphorylation also had significant up-regulation of genes associated with DNA damage (Bax: 1.71-fold, P=0.048; Mdm2: 1.65-fold, P=0.048; Ddb2: 1.89-fold, P=0.018; and Tp53: 1.87-fold, P=0.048, Figure 5C) at 48 hours post-SPECT, whereas those with no changes in phosphorylation had either significant down-regulation (Mdm2: 0.73-fold, P=0.003 and Ddb2: 0.60-fold, P=0.007) or no difference (Bax, Tp53, Atf6, and Bbc3) compared to baseline (Figure 5D). Taken together, these findings suggest that increased gene transcription occurs in tandem with increased phosphorylation of DNA damage marker proteins in a subset of patients undergoing SPECT MPI.
Evaluating Potential Factors Affecting Activation of the DNA Damage Response Pathways
To better define factors that might contribute to the variability of protein activation and gene expression, blood obtained from healthy volunteers with different age groups was exposed to 25 mGy of low dose radiation in vitro. Although not statistically significant, T-lymphocytes isolated in the young age group (20-25 years) appeared to have higher levels of phosphorylation compared to the middle age group (40-55 years), whose results were more comparable to those from our cohort of older individuals (mean age: 69.6 ± 10.5 years) (Supplemental Figure 6A-B). These findings were supported by a trend toward up-regulation in DNA damage response genes in the young age group compared to a lack of gene activation in the middle age group, which was consistent with our prior studies (Supplemental Figure 6C-D). Previous studies also have reported that certain subpopulations of T-lymphocytes may be more sensitive than others to radiation.25 To address whether this contributed to variability in the gene expression data, we next measured the effects of low dose radiation (e.g., 25 mGy) on the gene expression of central memory and naïve CD4+ and CD8+ T-lymphocytes. As shown in Supplemental Figure 7, although the overall trend showed a down-regulation of gene expression in all T-lymphocyte subpopulations 24 hours after exposure to low dose radiation, the level of expression was variable among different cell subtypes. Because our single cell PCR analyzed the expression of only a small set of cells, these differences in gene expression of T-lymphocyte subpopulations may have contributed to the inter-individual variability seen in our gene expression data.26
Discussion
SPECT MPI is a widely used non-invasive imaging technique for the diagnosis, risk stratification, and management of patients with CAD. Perfusion abnormalities are one of the earliest manifestations of ischemia, preceding the development of wall motion abnormalities and changes in the electrocardiogram detected by stress echocardiography and exercise treadmill testing, respectively.27 Patients with normal or low risk findings on SPECT have a less than 1% risk of developing a major adverse cardiac event at follow-up, which equates to a 98% negative predictive value for SPECT MPI. On the other hand, patients with high risk features such as extensive myocardial ischemia and myocardial perfusion defects involving multiple vascular territories have a predictive annual mortality of 3%. Not surprisingly, the diagnostic and prognostic accuracy of SPECT has led to a rapid increase of its use. Geographical variations in the utilization of SPECT MPI, however, have raised questions of over-utilization and concerns regarding unnecessary radiation exposure and its potential deleterious effects.28
In this prospective cohort study, we found that patients undergoing SPECT MPI had variable activation of the DNA damage response pathways. Most patients did not have significant changes in phosphorylation of DNA damage-marker proteins (i.e., H2AX, p53, and ATM), nor were there significant changes in mRNA expression of DNA damage response genes (i.e., Bax, Mdm2, Ddb2, and Tp53) detected in T-lymphocytes collected from patients post-SPECT compared with baseline. We did, however, notice a small cohort of patients who had increased protein phosphorylation post-SPECT, which was associated with up-regulation of genes involved in the DNA damage response pathways.
Exposure to high dose radiation results in cellular injury that activates the DNA damage response pathways, which has emerged as a surrogate marker of DNA damage. Previous studies have found a strong correlation between the number of DSBs and the degree of phosphorylation of proteins involved in the DNA damage response pathways.16, 17, 29 Recent studies have also shown that exposure of human blood cells to high doses of radiation (>100 mGy) significantly increases expression of genes involved in these pathways in a dose-dependent manner.21, 30 Similarly, other studies have found increased activation of these pathways in patients treated with high dose radiation therapy (>100 mGy).31, 32 Importantly, activation of the DNA damage response pathways has also been reported for low dose radiation (<100 mGy) exposure in vitro and in vivo. Separate studies have reported increased phosphorylation of H2AX in T-lymphocytes isolated from all adult patients after computed tomography (CT) imaging in a dose-dependent manner, but its effect on other DNA damage proteins remains largely unknown.4, 19, 20
Our results indicate a variable activation of DNA damage response pathways in patients undergoing SPECT MPI. Two-thirds of patients (n=29/45) had no change in protein phosphorylation, whereas a third of patients had evidence of increased phosphorylation in at least one protein marker of DNA damage. These findings are in stark contrast to results from another imaging modality, invasive X-ray angiography, in which all patients (n=12/12) undergoing cardiac catheterization had increased levels of protein phosphorylation in at least one of the protein markers of DNA damage that we tested.
In addition to reports showing increased protein phosphorylation after low dose radiation exposure, changes in gene expression have been reported in vitro.24 Using a DNA microarray, Nosel et al. found both up- and down-regulation of genes involved in the DNA damage response pathways after exposure to 25-500 mGy of radiation in vitro. Interestingly, at 25 mGy, only genes involved in the regulation of cell death processes but not genes in cell cycle regulation were induced; this is consistent with both our in vivo and in vitro data, although differences in gene expression between baseline and post-SPECT were not significant. In fact, only 6 genes (Bax, Tp53, Ddb2, Mdm2, Atf6, and Bbc3) were found to be the most responsive in T-lymphocytes after exposure to low dose radiation. Similar to changes in protein phosphorylation, we observed more consistent up-regulation of genes in patients undergoing invasive X-ray angiography. This is in contrast to those undergoing SPECT MPI, for which most patients showed down-regulation or no change in mRNA expression of these genes within 48 hours post-exposure, once again demonstrating that SPECT produces a variable activation of the DNA damage response pathways. It should be noted that compared to either invasive X-ray angiography or SPECT MPI, we did not observe any changes in either protein phosphorylation or gene expression in patients undergoing echocardiography. These results argue against the possibility that the variable activation of the DNA damage response pathways seen in patients undergoing SPECT is due to natural circadian transcriptional and post-transcriptional changes that may be unrelated to ionizing radiation.
While this study does not explain the discrepancies observed in the extent of protein and gene activation among these patients undergoing SPECT MPI versus invasive X-ray angiography, one plausible explanation for the discrepancies may lie in the differences in the amount or frequency of dosing between these imaging modalities. Patients undergoing CT and invasive X-ray angiography are exposed to a continuous burst of radiation over a short period of time (<30 minutes), resulting in a “single” exposure of ~5-10 mSv. Patients undergoing SPECT MPI, on the other hand, have a nuclear tracer delivered to them intravenously, in two divided doses (~3 mSv followed by ~8 mSv) separated by 90 minutes in between. It is possible that some patients undergoing SPECT MPI did not reach the threshold dose that results in DNA damage as a result of fractionated dosing.
The marked variation in response to low dose radiation in patients undergoing SPECT MPI suggests that a subset of patients may be more sensitive to radiation. This could mean that while the majority of patients experience no changes in protein markers of DNA damage at these relatively low radiation doses, a subset of patients do suffer significant changes. Interestingly, this more sensitive subset of patients with increased protein phosphorylation also had up-regulation of DNA damage response genes, suggesting an ongoing activation of these pathways. Previous studies have demonstrated individual variability in radiation sensitivity at the level of gene expression.33, 34 Using genetic mapping, Smirnov et al. identified individual polymorphisms in regulators of gene expression that contributed to individual differences in radiation sensitivity.33 However, questions as to whether there is any genetic predisposition among certain patients affecting the cellular response to SPECT MPI, or whether these changes at both gene and protein levels reflect persistent DNA damage, are beyond the scope of the present study.
There are several limitations to this study. First, we did not measure gene and protein changes at multiple time points in vivo in all patients; thus, it is possible that we might have missed the exact peak changes in response. However, our initial studies in a subset of patients showed that changes in protein phosphorylation and gene expression are still detectable at 30 minutes and 24-48 hours, respectively. Second, SPECT may produce changes in protein phosphorylation that are undetectable by flow cytometry. The advantage of flow cytometry is its ability to simultaneously screen thousands of cells for multiple markers of DNA damage. Although this technique is potentially less sensitive than immunohistochemistry, we found no significant differences in phosphorylation changes as measured by flow cytometry versus immunohistochemistry in a subset of patients. Third, we did not directly measure DNA damage, and it is possible that patients who have greater activation of these pathways might not have more DSBs, but were somehow able to better respond to small doses of radiation. Thus the persistence of induced DNA damage is unknown, and there are no available techniques to directly quantify a small number of DSBs. Lastly, a demonstration of variable DNA damage pathway activation induced by SPECT does not necessarily equate to increased cancer risk. Our results were determined in peripheral blood lymphocytes, which are one of the most radiosensitive cell types in the body, but nevertheless may not directly lead to cancer.35 Rather, our results represent a “snapshot” of the cellular response to SPECT-induced radiation damage. Additionally, the small increased risk in cancer induction due to radiation is particularly difficult to detect as it is neither differentiable nor predictable for individual patients. This is further complicated by the omnipresent background radiation and also the fact that inherent (baseline) risk of cancer is much higher than the potential risk of radiation-induced cancer.3, 35, 36 Hence, determining specific cancer risks is beyond the scope of this study and is likely not feasible using any existing strategies.
Taken together, our results show that the majority of patients undergoing SPECT MPI did not show an activation of the DNA damage response pathways. However, a small minority of patients did experience increased phosphorylation, which is associated with up-regulation of genes involved in the DNA damage response pathways. The variable activation induced by SPECT differed from that seen in invasive X-ray angiography, which produced a uniform activation pattern; it was also different compared to echocardiography, which showed an absence of activation of the DNA damage response pathways. It is reasonable to conclude that although the risk of low dose radiation from SPECT MPI remains a valid concern, most patients who undergo imaging for diagnosis and risk stratification are often of advanced age and may benefit more from cardiac imaging procedures relative to the potential harms of radiation-based imaging during the remainder of their expected life span.36 However, our results also show that the risk of low-dose radiation is not zero, and therefore it should continue to be standard practice to perform all imaging studies at the lowest possible radiation dose without compromising diagnostic image quality.
Supplementary Material
Acknowledgemtns
We would like to thank the Stanford Functional Genomics Facility and Stanford FACS Facility for their continued support and insight. We thank Jarrett Rosenberg, PhD for his assistance with the statistical analysis. We would like to thank Joseph Gold, PhD and Blake Wu for their review and contribution to the manuscript.
Sources of Funding
American Heart Association 10SDG4280129 grant (PKN), National Institutes of Health R01 EB009689 (JCW), and the Stanford Cardiovascular Institute (PKN, JCW)
Footnotes
Disclosures
None.
References
- 1.Chen J, Einstein AJ, Fazel R, Krumholz HM, Wang Y, Ross JS, Ting HH, Shah ND, Nasir K, Nallamothu BK. Cumulative exposure to ionizing radiation from diagnostic and therapeutic cardiac imaging procedures: a population-based analysis. J Am Coll Cardiol. 2010;56:702–711. doi: 10.1016/j.jacc.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw LJ, Marwick TH, Zoghbi WA, Hundley WG, Kramer CM, Achenbach S, Dilsizian V, Kern MJ, Chandrashekhar Y, Narula J. Why all the focus on cardiac imaging? JACC. Cardiovasc Imaging. 2010;3:789–794. doi: 10.1016/j.jcmg.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Tubiana M, Feinendegen LE, Yang C, Kaminski JM. The linear no-threshold relationship is inconsistent with radiation biologic and experimental data. Radiology. 2009;251:13–22. doi: 10.1148/radiol.2511080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lobrich M, Rief N, Kuhne M, Heckmann M, Fleckenstein J, Rube C, Uder M. In vivo formation and repair of DNA double-strand breaks after computed tomography examinations. Proc Natl Acad Sci U S A. 2005;102:8984–8989. doi: 10.1073/pnas.0501895102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breckow J. Linear-no-threshold is a radiation-protection standard rather than a mechanistic effect model. Radiat Environ Biophys. 2006;44:257–260. doi: 10.1007/s00411-006-0030-y. [DOI] [PubMed] [Google Scholar]
- 6.Brenner DJ. Radiation risks potentially associated with low-dose CT screening of adult smokers for lung cancer. Radiology. 2004;231:440–445. doi: 10.1148/radiol.2312030880. [DOI] [PubMed] [Google Scholar]
- 7.Tubiana M, Aurengo A, Averbeck D, Masse R. Recent reports on the effect of low doses of ionizing radiation and its dose-effect relationship. Radiat Environ Biophys. 2006;44:245–251. doi: 10.1007/s00411-006-0032-9. [DOI] [PubMed] [Google Scholar]
- 8.Feinendegen LE, Pollycove M. Biologic responses to low doses of ionizing radiation: detriment versus hormesis. Part 1. Dose responses of cells and tissues. J Nucl Med. 2001;42:17N–27N. [PubMed] [Google Scholar]
- 9.Feinendegen LE. Evidence for beneficial low level radiation effects and radiation hormesis. Br J Radiol. 2005;78:3–7. doi: 10.1259/bjr/63353075. [DOI] [PubMed] [Google Scholar]
- 10.Marchetti F, Coleman MA, Jones IM, Wyrobek AJ. Candidate protein biodosimeters of human exposure to ionizing radiation. Int J Radiat Biol. 2006;82:605–639. doi: 10.1080/09553000600930103. [DOI] [PubMed] [Google Scholar]
- 11.Nur EKA, Li TK, Zhang A, Qi H, Hars ES, Liu LF. Single-stranded DNA induces ataxia telangiectasia mutant (ATM)/p53-dependent DNA damage and apoptotic signals. J Biol Chem. 2003;278:12475–12481. doi: 10.1074/jbc.M212915200. [DOI] [PubMed] [Google Scholar]
- 12.Mattsson S, Johansson L, Leide-Svegborn S, Liniecki J, Nosske D, Riklund K, Stabin M, Taylor D. Current activities in the ICRP concerning estimation of radiation doses to patients from radiopharmaceuticals for diagnostic use. J Phys: Conf Ser. 2011:317. [Google Scholar]
- 13.Einstein AJ, Moser KW, Thompson RC, Cerqueira MD, Henzlova MJ. Radiation dose to patients from cardiac diagnostic imaging. Circulation. 2007;116:1290–1305. doi: 10.1161/CIRCULATIONAHA.107.688101. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez-Freire V, Ebert AD, Kalisky T, Quake SR, Wu JC. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nat Protoc. 2012;7:829–838. doi: 10.1038/nprot.2012.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Beels L, Bacher K, De Wolf D, Werbrouck J, Thierens H. Gamma-H2AX foci as a biomarker for patient X-ray exposure in pediatric cardiac catheterization: are we underestimating radiation risks? Circulation. 2009;120:1903–1909. doi: 10.1161/CIRCULATIONAHA.109.880385. [DOI] [PubMed] [Google Scholar]
- 17.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 18.Wittlinger M, Grabenbauer GG, Sprung CN, Sauer R, Distel LV. Time and dose-dependent activation of p53 serine 15 phosphorylation among cell lines with different radiation sensitivity. Int J Radiat Biol. 2007;83:245–257. doi: 10.1080/09553000701275432. [DOI] [PubMed] [Google Scholar]
- 19.Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci U S A. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothkamm K, Balroop S, Shekhdar J, Fernie P, Goh V. Leukocyte DNA damage after multi-detector row CT: a quantitative biomarker of low-level radiation exposure. Radiology. 2007;242:244–251. doi: 10.1148/radiol.2421060171. [DOI] [PubMed] [Google Scholar]
- 21.Turtoi A, Brown I, Oskamp D, Schneeweiss FH. Early gene expression in human lymphocytes after gamma-irradiation-a genetic pattern with potential for biodosimetry. Int J Radiat Biol. 2008;84:375–387. doi: 10.1080/09553000802029886. [DOI] [PubMed] [Google Scholar]
- 22.Gruel G, Voisin P, Vaurijoux A, Roch-Lefevre S, Gregoire E, Maltere P, Petat C, Gidrol X, Roy L. Broad modulation of gene expression in CD4+ lymphocyte subpopulations in response to low doses of ionizing radiation. Radiat Res. 2008;170:335–344. doi: 10.1667/RR1147.1. [DOI] [PubMed] [Google Scholar]
- 23.Mori M, Benotmane MA, Tirone I, Hooghe-Peters EL, Desaintes C. Transcriptional response to ionizing radiation in lymphocyte subsets. Cell Mol Life Sci. 2005;62:1489–1501. doi: 10.1007/s00018-005-5086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nosel I, Vaurijoux A, Barquinero JF, Gruel G. Characterization of gene expression profiles at low and very low doses of ionizing radiation. DNA Repair (Amst) 2013;12:508–517. doi: 10.1016/j.dnarep.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 25.Schmitz A, Bayer J, Dechamps N, Thomas G. Intrinsic susceptibility to radiation-induced apoptosis of human lymphocyte subpopulations. Int J Radiat Oncol Biol Phys. 2003;57:769–778. doi: 10.1016/s0360-3016(03)00637-0. [DOI] [PubMed] [Google Scholar]
- 26.Ponnaiya B, Amundson SA, Ghandhi SA, Smilenov LB, Geard CR, Buonanno M, Brenner DJ. Single-cell responses to ionizing radiation. Radiat Environ Biophys. 2013;52:523–530. doi: 10.1007/s00411-013-0488-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klocke FJ, Baird MG, Lorell BH, Bateman TM, Messer JV, Berman DS, O'Gara PT, Carabello BA, Russell RO, Jr., Cerqueira MD, St John Sutton MG, DeMaria AN, Udelson JE, Kennedy JW, Verani MS, Williams KA, Antman EM, Smith SC, Jr., Alpert JS, Gregoratos G, Anderson JL, Hiratzka LF, Faxon DP, Hunt SA, Fuster V, Jacobs AK, Gibbons RJ, Russell RO. Acc/aha/asnc guidelines for the clinical use of cardiac radionuclide imaging--executive summary: A report of the american college of cardiology/american heart association task force on practice guidelines (acc/aha/asnc committee to revise the 1995 guidelines for the clinical use of cardiac radionuclide imaging). Circulation. 2003;108:1404–1418. doi: 10.1161/01.CIR.0000080946.42225.4D. [DOI] [PubMed] [Google Scholar]
- 28.Parker L, Levin DC, Frangos A, Rao VM. Geographic variation in the utilization of noninvasive diagnostic imaging: National medicare data, 1998-2007. AJR Am J Roentgenol. 2010;194:1034–1039. doi: 10.2214/AJR.09.3528. [DOI] [PubMed] [Google Scholar]
- 29.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 30.Amundson SA, Do KT, Shahab S, Bittner M, Meltzer P, Trent J, Fornace AJ., Jr. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res. 2000;154:342–346. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 31.Sak A, Grehl S, Erichsen P, Engelhard M, Grannass A, Levegrun S, Pottgen C, Groneberg M, Stuschke M. Gamma-H2AX foci formation in peripheral blood lymphocytes of tumor patients after local radiotherapy to different sites of the body: dependence on the dose-distribution, irradiated site and time from start of treatment. Int J Radiat Biol. 2007;83:639–652. doi: 10.1080/09553000701596118. [DOI] [PubMed] [Google Scholar]
- 32.Amundson SA, Grace MB, McLeland CB, Epperly MW, Yeager A, Zhan Q, Greenberger JS, Fornace AJ., Jr. Human in vivo radiation-induced biomarkers: gene expression changes in radiotherapy patients. Cancer Res. 2004;64:6368–6371. doi: 10.1158/0008-5472.CAN-04-1883. [DOI] [PubMed] [Google Scholar]
- 33.Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG. Genetic analysis of radiation-induced changes in human gene expression. Nature. 2009;459:587–591. doi: 10.1038/nature07940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smirnov DA, Brady L, Halasa K, Morley M, Solomon S, Cheung VG. Genetic variation in radiation-induced cell death. Genome Res. 2012;22:332–339. doi: 10.1101/gr.122044.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerber TC, Gibbons RJ. Weighing the risks and benefits of cardiac imaging with ionizing radiation. JACC Cardiovasc Imaging. 2010;3:528–535. doi: 10.1016/j.jcmg.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Gerber TC, Carr JJ, Arai AE, Dixon RL, Ferrari VA, Gomes AS, Heller GV, McCollough CH, McNitt-Gray MF, Mettler FA, Mieres JH, Morin RL, Yester MV. Ionizing radiation in cardiac imaging: a science advisory from the American Heart Association Committee on Cardiac Imaging of the Council on Clinical Cardiology and Committee on Cardiovascular Imaging and Intervention of the Council on Cardiovascular Radiology and Intervention. Circulation. 2009;119:1056–1065. doi: 10.1161/CIRCULATIONAHA.108.191650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.