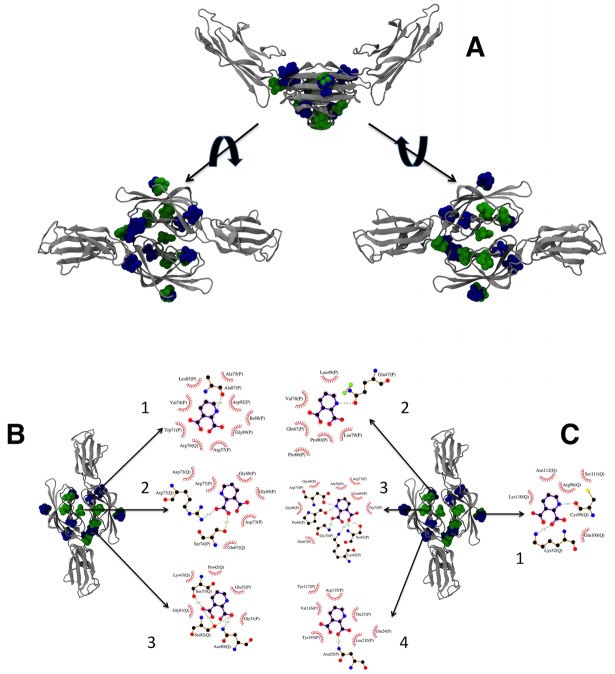
Fig 8. Docking for VC1-QUIN.
In A, the structures of the 20 highest scoring conformers of QUIN obtained by docking on all surfaces of VC1 human (green) and rat (blue) RAGE domains. The topmost figure shows the V-shaped dimer in silver ribbons, with the QUIN molecules in spheres. The left structure shows the dimer from the top, and the right structure, from the bottom. In B, the three binding sites that involve residues from both monomers are shown (labels 1, 2, and 3), together with the details of the interacting residues for the best pose of each site using LIGPLOT+ [74] with the default parameters. In C, the four binding sites that only involve residues from one monomer are shown (labels 1 through 4), together with details of the interacting residues for the best pose of each site using LIGPLOT+ [74] with the default parameters. The 3D structures were prepared with VMD 1.9.1 [75]. In panels B and C, residue names and numbers are followed by the chain identifier (P or Q). Hydrogen bonds are indicated by green dashed lines with the distance between heavy atoms, and van der Waals contacts are indicated by red arcs with short lines. Residue numbers correspond to the full chain numbering from rat or human.