Figure 1.
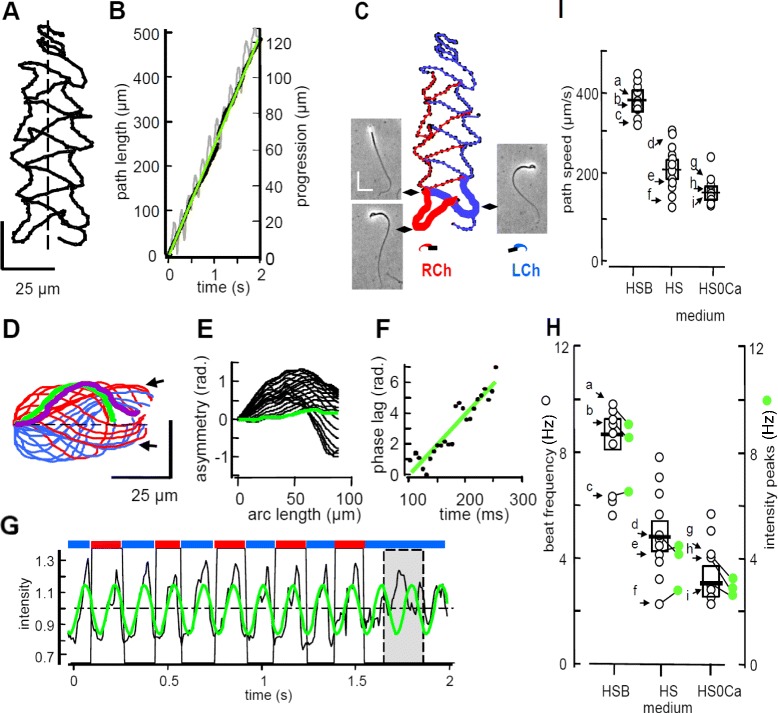
Linear mean swimming trajectories comprise regularly alternating changes in sperm orientation synchronized with the flagellar beat cycle. (A) Tracking a typical free-swimming sperm. Duration 2 s, sampling 150 frames per s (fps), medium HSB. Dashed line approximates mean swimming trajectory. (B) Time course of progress along swimming path. Point-to-point movement of head centroid defines path (black). Best fit linear regression lines (green) indicate mean path speed and progression (gray, linear distance from initial position). (C) Two initial segments of swimming track (heavy lines), flanking cell images at indicated track locations. Red, blue indicate cell orientation with right cheek (RCh), left cheek (LCh) facing bottom surface. Scale bar 25 μm. (D) Aligned flagellar traces (75 fps sampling) during initial RCh (red) and LCh (blue) segments. Initial trace green; final trace purple. Arrows indicate linear waveforms observed near RCh-to-LCh transition. (E) Flagellar waveform asymmetry from running averages of tangent angles (shear angles, in radians) for traces in D. Final, time-averaged shear angle is green. (F) Flagellar beat frequency (6.31 Hz) reported by phase lag of sine curves fitted to traces in D. Phase changes by 2π radians each beat cycle. (G) Normalized intensity from 7 × 7 pixel region-of-interest (ROI) enclosing the centroid (black) oscillates regularly (green). Red and blue bars (top) mark RCh, LCh orientation. Dashed box encloses interval when another motile cell interferes with intensity signal. (H) Flagellar beat frequency (black) correlates with frequency of ROI intensity peaks (green). Lines connect frequency pairs from same cell. Arrows and letters (a to i) indicate cells also analyzed as follows. (I) Path speeds for cells in resting medium (HS, n = 12 cells), activating HCO3−-fortified medium (HSB, n = 13 cells), or Ca2+-deficient media (HS0Ca, n = 12 cells). Note rank order within each medium is the same as in panel H.
