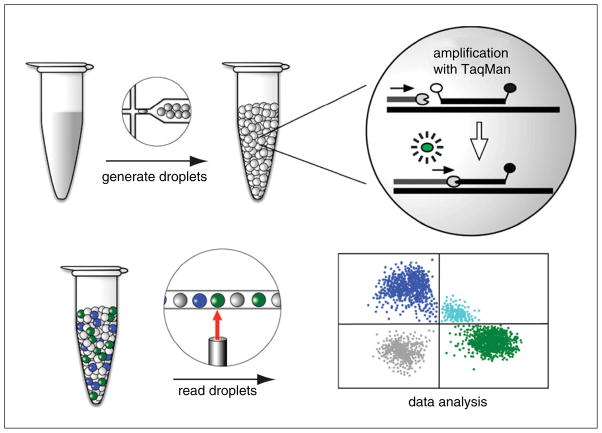
Figure 7.24.1.
Overview of digital droplet PCR workflow. The PCR mix is partitioned into thousands of nanoliter-sized droplets using specialized oil and microfluidics technology. The droplets are subjected to thermal cycling in a standard PCR machine. A PCR reaction only occurs in those droplets carrying the target DNA. PCR products are detected by a TaqMan assay. Briefly, the TaqMan probe hybridizes to an internal site of the PCR product. The probe has a 5′ fluorophore whose fluorescence is quenched by a 3′ quencher. Once the DNA polymerase reaches the probe during the extension step, the probe is cleaved by the polymerase’s 5′ to 3′ exonuclease activity. This action liberates the fluorophore from the quencher so that the fluorescence can be detected when excited by the appropriate wavelength of light. Droplets are read one by one using a droplet reader for fluorescence in each of two channels (FAM in blue and VIC in green). The data are displayed in a 2-D plot of positive droplets in the FAM and VIC channels, double-positive (aqua), and double-negative (gray). See text for further details.