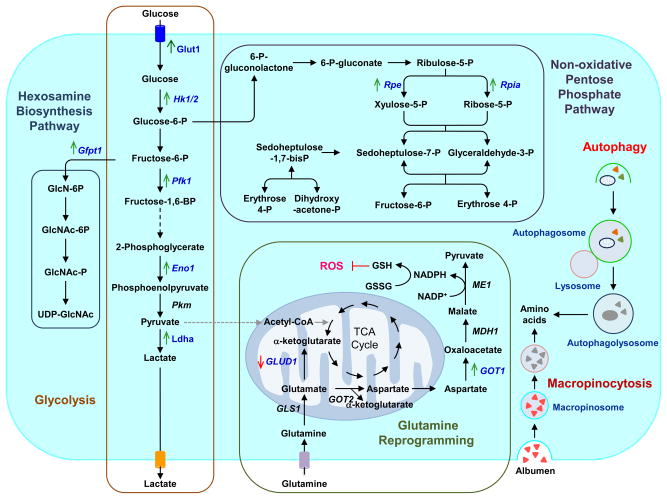
Figure 6. Ras-driven alterations in metabolism.
RAS-mutant cancer cells are characterized by increased macropinocytosis and uptake of albumen, leading to lysosomal degradation and release of amino acids. RAS-mutant cancer cells also exhibit altered autophagy, leading to degradation of organelles and proteins, and to production of amino acids and other components that support metabolism. Oncogenic K-Ras directs glucose metabolism into biosynthetic pathways in PDAC by upregulating many key enzymes in glycolysis. Oncogenic K-Ras induces nonoxidative PPP flux to fuel increased nucleic acid biosynthesis and activates the hexosamine biosynthesis and glycosylation pathways. PDAC cells also utilize a non-canonical pathway to process glutamine and use it to maintain redox status and support growth. Blue text indicates Ras-dependent gene and/or protein expression, with arrows indicating increased (green) or decreased (red) expression. Enzymes are indicated in italics. Abbreviations used are: Glut1, glucose transporter 1; Hk 1/2, hexokinase 1/2; Pfk1, phosphofructokinase 1; Eno1, enolase 1; Pkm, pyruvate kinase; Ldha, lactate dehydrogenase A; Gfpt1, glucosamine-fructose-6-phosphate aminotransferase-1; GlcN, glucosamine; GlcNAc, N-acetylglucosamine; Rpe, ribulose-5-phosphate-3-epimerase; Rpia, ribulose-5-phosphate isomerase; GLUD1, glutamate dehydrogenase 1; GLS1, glutaminase 1; GOT1/2, aspartate transaminase 1/2; MDH1, malate dehydrogenase 1; ME1, malic enzyme; GSH, glutathione; GSSG, glutathione disulfide; ROS, reactive oxygen species.