Abstract
Six proteins, ORC1-6, make up the origin recognition complex (ORC) that initiates licensing of DNA replication origins. We have previously reported that subunit ORC2 is localized between the separating maternal chromosomes at anaphase II just after fertilization and is present in zygotic pronuclei at G1. Here, we found that ORC1, 3, and 5 all localize between the chromosomes at anaphase II, but could not be detected in zygotic G1. ORC6 localized to the periphery of the nucleoli at all zygotic stages. We identified an unexpected potential role for ORC4 in polar body formation. We found that in both female meiotic divisions, ORC4 surrounds the set of chromosomes, as a sphere-like structure, that will eventually be discarded in the polar bodies, but not the chromosomes that segregate into the oocyte. None of the other five ORC proteins are involved in this structure. In Zygotic G1, ORC4 surrounds the nuclei of the polar bodies, but was not detectable in the pronuclei. When the zygote entered mitosis ORC4 was only detected in the polar body. However, ORC4 appeared on both sets of separating chromosomes at telophase. At this point, the ORC4 that was in the polar body also migrated into the nuclei, suggesting that ORC4 or an associated protein is modified during the first embryonic cell cycle to allow it to bind DNA. Our results suggest that ORC4 may help identify the chromosomes that are destined to be expelled in the polar body, and may play a role in polar body extrusion. extrusion. ORC4 surrounds the chromatin that will be extruded in the polar body in both female meiotic divisions, then makes a transition from the cytoplasm to the chromosomes at zygotic anaphase, suggesting multiple roles for this replication licensing protein.
Keywords: ORC1, ORC2, ORC3, ORC4, ORC5, ORC6, polar body extrusion, DNA licensing, mouse zygote
Introduction
Origins are marked by ORC (origin recognition complex) proteins during the M-G1 transition to ensure all DNA is only replicated once during S-phase [DePamphilis, 2005; Krude, 2006; Takeda and Dutta, 2005; Thomae et al., 2008]. This is the first step of a process called licensing that serves as a bookkeeping mechanism for the cell to monitor its progress in duplicating the chromosomes. The ORC is made up of six proteins, ORC1-6 ranging from 95 to 29 kD (NCBI database). The ORC complex proteins), which are found in all types of cells. from bacteria, in which DnaA is structurally and functionally homologous to ORC to mammals and are part of a group of proteins termed replication initiators [Scholefield et al., 2011]. The six ORC proteins were originally discovered in yeast and named according to size, with ORC1 being the largest, and the subunits in other species names according to the homology with their yeast counterparts [Li and Stillman, 2012]. More studies have been done on human ORC than mouse, but the close homology between these two species suggests that similar mechanisms are involved in both [Kneissl et al., 2003; Springer et al., 1999a; Springer et al., 1999b]. Previous models for human ORC proposed that in actively dividing somatic mammalian cells, ORC2-5 remain associated with DNA replication origins throughout the cell cycle while ORC1 is recruited to the ORC complex only during G1 [DePamphilis, 2005; Dhar et al., 2001; Thomae et al., 2008; Vashee et al., 2001]. However, more recent work has suggested that it is possible all the ORC proteins are recruited to the origins during each cycle [Ghosh et al., 2011]. Moreover, the DNA binding patterns of ORC to DNA vary during differentiation [Nordman and Orr-Weaver, 2012].
Licensing patterns during fertilization and subsequent early embryo development are particularly unique. Sonneville et al. [Sonneville et al., 2012] demonstrated that ORC2, one of the first ORC proteins to bind to naïve DNA origins in the C. elegans zygote, binds DNA only transiently, long enough to recruit the MCM7 helicases that represent the final step of licensing. We previously demonstrated that the maternal DNA is likely to be licensed before fertilization but the paternal DNA is licensed de novo in the embryo [Ortega et al., 2012]. We also identified unexpected roles for ORC2 in the first cell cycle. We demonstrated that ORC2 binds to the DNA in both pronuclei in late zygotic G1 as expected for its role in licensing. However, just after fertilization in anaphase II, ORC2 localized between the two separating sets of maternal chromosomes. While the original role of ORC proteins was to mark DNA replication origins, it has now been suggested that they also play more diverse roles that coordinate DNA replication with other cell division events [Sasaki and Gilbert, 2007; Scholefield et al., 2011].
Here, we examined the roles of the other five ORC proteins in the first cell cycle. We found that ORC1, ORC3, and ORC5 had similar patterns of localization to ORC2 at anaphase II, but ORC6 and ORC4 exhibited surprisingly different patterns. In particular, the data suggest that ORC4 may play a role in polar body extrusion.
Methods
MATERIAL AND METHODS
Animals
Eight-week old B6D2F1 (C57BL/6N X DBA/2) mice were used. Mice were obtained from National Cancer Institute (Raleigh, NC), and used at 8 – 12 weeks of age. Animal care and experimental protocols for handling and the treatment procedures were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Hawai’i.
Preparation of Spermatozoa
Spermatozoa were obtained from B6D2F1 male mice (8-12 week old). A caudal epididymis was extracted, suspended in HEPES-CZB, and incubated for 45 minutes at 37°C under 5% CO2 in air to allow the spermatozoa to disperse in the medium. The spermatozoa suspension for Intracytoplasmic sperm injection (ICSI) was mixed with PVP-saline (0.9% NaCl containing 12% w/v polyvinyl pyrolidone [360 kDa; ICN Biochemicals])
Preparation of Oocytes
To obtain mature metaphase II (MII) oocytes, mature female, 8 – 10 week-old, were induced to superovulate by injections of 5 IU PMSG and 5 IU hCG given 48h apart. Oviducts were removed 14 -15 hours after the injections of hCG and placed in mineral oil. The cumulus cells were released from the oviducts into 0.1% bovine testicular hyaluronidase/CZB medium for 10 minutes to disperse cumulus cells. The cumulus-free oocytes were washed and kept in CZB at 37° C under 5% CO2 in air. These oocytes were used for immunocytochemistry (ICC) and ICSI.
Intracytoplasmic Sperm Injection (ICSI)
ICSI was carried out as described by Kimura and Yanagimachi [Kimura and Yanagimachi, 1995]. ICSI was performed using Eppendorf Micromanipulators (Micromanipulator TransferMan, Eppendorf, Germany) with a piezoelectric actuator (PMM Controller, model PMAS-CT150; Prime Tech, Tsukuba, Japan). A single sperm head was sucked, tail first, into the injection pipette and moved back and forth until the head-mid piece junction (the neck) was at the opening of the injection pipette. The head was separated from the mid piece by applying one or more piezo pulses. An oocyte was held to the holding pipette at the 6 or 9 o'clock position, and the sperm head was redrawn into the pipette and injected immediately into an oocyte. After discarding the mid piece and tail, the sperm head was redrawn into the pipette and injected immediately into an oocyte. After ICSI, oocytes were cultured in CZB at 37° C under 5% CO2 in air.
Preparation immature oocytes
To collect immature metaphase I and anaphase I oocytes, females were induced to super-ovulate by injections of 5 IU of PMSG. Ovaries were removed 48 hours after PMSG injections and placed in HEPES-CZB in a petri dish. The biggest follicles were broken to release germinal vesicle stage (GV) oocytes. GV oocytes with surrounded cumulus cells were placed in CZB drops under mineral oil and cultured for 2 hours. After 2 hours, cumulus cells were removed by pipetting. The oocytes that underwent germinal vesicle breakdown were collected and cultured for 4 and 9 hours to reach metaphase I and anaphase I, respectively. Oocytes were then used for ICC.
Antibodies
Polyclonal goat anti-ORC1 (N-17, catalog no. sc-13231; Santa Cruz Biotechnology, Santa Cruz, CA ), polyclonal goat anti-ORC2 (C-18, catalog no. sc-13238; Santa Cruz Biotechnology, Santa Cruz, CA), Polyclonal goat anti-ORC3 (K-17, catalog no. sc-21862; Santa Cruz Biotechnology, Santa Cruz, CA ), Polyclonal goat anti-ORC4 (C-15, catalog no. sc-19726; Santa Cruz Biotechnology, Santa Cruz, CA ), Polyclonal goat anti-ORC5 (T-15, catalog no. sc-19728; Santa Cruz Biotechnology, Santa Cruz, CA ), Polyclonal goat anti-ORC6 (2679C2b, catalog no. sc-81646; Santa Cruz Biotechnology, Santa Cruz, CA ) and monoclonal mouse-MCM7 (H-5, catalog no. sc-374403; Santa Cruz Biotechnology) primary antibodies were used. The Secondary antibodies included Alexa Fluor 488 rabbit anti-goat and Alexa Fluor 546 rabbit anti-mouse (Invitrogen, Grand Island, NY).
Immunocytochemistry (ICC)
Embryos were cultured in CZB until they reached the desired stage after ICSI, and fixed in 2% paraformaldehyde for 30 minutes at room temperature. After fixing, cells were washed three times with 0.1% Tween/PBS (PBSw) for 10 minutes. Cells were permeabilized in 0.5% Triton X-100 for 15 minutes, after that the cells were washed three times with PBSw contained 10% volume of 5% BSA. Cells were blocked with 5% BSA for 1 h at room temperature, then incubated in primary antibody at 1:50 dilution overnight at 4 o C. Again, the cells were washed three times with PBSw contained 10% volume of 5% BSA, then incubated in secondary antibody at 1:1500 dilutions at room temperature for 45 minutes. Cells were again washed three times with PBSw contained 10% volume of 5% BSA then mounted with ProLong Gold antifade reagent with DAPI (catalog no. P-36931; Invitrogen). ICC was analyzed with an FV1000-IX81 confocal microscope from Olympus using Fluoview v. 2.1 software. For each stage of embryonic development, at least 20 embryos were examined by ICC, and the results were only reported if they were consistent in all embryos.
Results
ORC1, ORC3, ORC5 and ORC6 localize between the chromosomes at anaphase II similar to ORC2
We recently examined the expression pattern of ORC2 during the first two cell cycles of the mouse embryo. ORC2 localizes at the area between the separating chromosomes in anaphase II just after fertilization, then moves into the pronuclei where it becomes resistant to salt extraction, indicative of DNA binding [Ortega et al., 2012]. To complete the analysis of the expression patterns of all six ORC proteins, we examined ORC1, and ORC3-6 during the first cell cycle of the mouse zygote. We found that ORC1, ORC3, and ORC5 behave similarly to ORC2 in that they localized between the separating chromosomes at anaphase II (Fig. 1, Suppl. Fig. 1) and to the spindle poles zygotic metaphase (Suppl Fig. 2). ORC1 surrounded the chromosomes at zygotic anaphase, but did not significantly overlap with the chromosomes (Suppl. Fig. 3), just as we had previously seen for ORC2 [Ortega et al., 2012]. This suggests that ORC1, ORC2, ORC3 and ORC5 may play structural roles during mitosis. ORC1, ORC3 and ORC5 could not be detected in the zygotoc pronuclei, however (data not shown). This differs from ORC2, which is easily detectable in the zygotic pronuclei at all stages [Ortega et al., 2012]. ORC6, on the other hand, was found only in the periphery of the nucleoli at all stages between zygotic G1 and 2-Cell G1 (Figs. 2B and J, Suppl. Fig. 4). During zygotic telephase, ORC6 was present around the nucleoli of the polar body, but was not detectable on the chromosomes (Fig. 2F).
Figure 1. ORC1-3 and ORC5 are adjacent to the chromosomes at anaphase II.
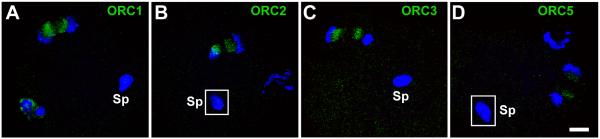
ICSI generated zygotes were fixed and stained for ORC1 (A), ORC2 (B), ORC3 (C) and ORC5 (D) and visualized by confocal microscopy. All four ORC subunits localize to the area between separating maternal chromosomes in anaphase II. The decondensing sperm nucleus had no visible staining in any oocyte. Insets in (B) and (D) represent areas in different confocal planes than the maternal chromatin. Sp, decondensing sperm nucleus. All images are shown at the same magnification, bar = 10 μm.
Figure 2. ORC6 in the first zygotic cell cycle.
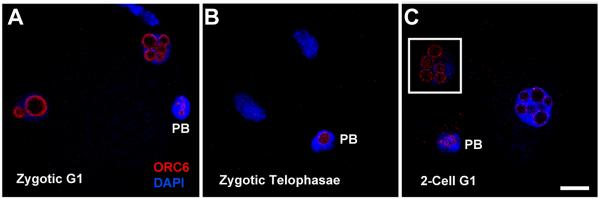
ICSI generated zygotes were fixed and stained for ORC6 at zygotic G1 (A), zygotic telophase (B), and 2-cell G1 (C) and visualized by confocal microscopy. Inset in (C) is the area in different confocal plane from the main image that contained the second nucleus. PB, second polar body. All images are shown at the same magnification, bar = 10 μm. An expanded version of this figure is shown in Suppl. Fig. 4.
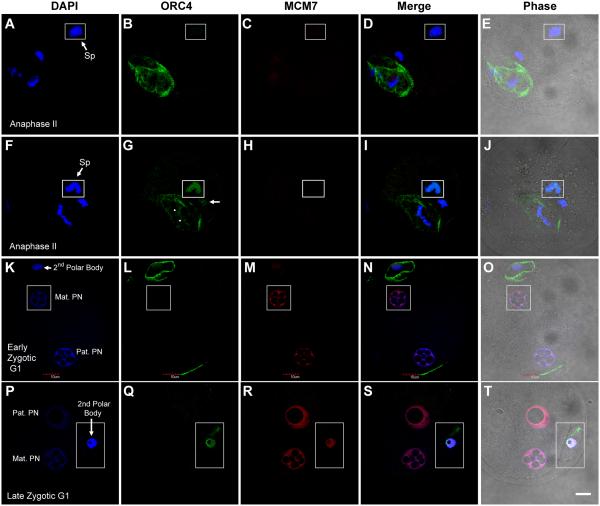
ORC4 surrounds the polar body chromatin at anaphase II
We were surprised to find that ORC4 behaves in a very different manner than the other ORC proteins. At anaphase II, ORC4 formed a ring, as visualized by confocal microscopy, around the set of chromosomes that would eventually segregate into the second polar body (Fig. 3B). Two different antibodies gave us the same result. This was a consistent finding in every zygote at anaphase II for which ORC4 staining was clear. ORC4 always surrounded the chromosomes that were nearest the zygotic membrane, and therefore presumed to be those destined to segregate into the second polar body. This assumption is supported by the fact that in zygotic G1 after the polar body had been extruded, ORC4 was localized to the nuclear periphery of the polar body nucleus (Fig. 3L). ORC4 was not detectable in the pronuclei in the zygote at this stage. The decondensing sperm chromatin in the zygote after fertilization was initially not associated with ORC4 (Fig. 3B), but as decondensation progressed, ORC4 was found to be associated with sperm chromatin in a 3/12 zygotes (Fig. 3G, inset). At this point, ORC4 also bound to both sets of maternal chromatin, the one that would contribute to the maternal proncleus (Fig. 3G, arrow) and one that would be segregated into the polar body (Fig. 3G, arrowheads). The other two examples of ORC4 binding to sperm and maternal chromatin during anaphase II are shown in Suppl. Fig. 5. The fact that so few zygotes had ORC4 bound to the sperm chromatin suggested that this binding seemed to be transient. Neither of the two pronuclei at zygotic G1 had detectable ORC4 (Fig. 3L).
Figure 3. ORC4 surrounds the polar body chromatin during anaphase II.
ICSI generated zygotes were fixed and stained for ORC4 at early anaphase II (A-J), early zygotic G1 (K-O) and late zygotic G1 (P-T), and visualized by confocal microscopy. ORC4 surrounds the chromatin that is closest to the oolemma (B and G), and the nucleus of the polar body at G1 (L). ORC4 was also visible in the decondensing sperm nucleus (G, inset) but not in a sperm that was less decondensed (B). ORC4 translocates to the nucleus of the second polar body in late G1 (Q). All insets show areas from different confocal planes than the rest of the figure. All images are shown at the same magnification, bar = 10 μm.
ORC4 also surrounds first polar body chromatin
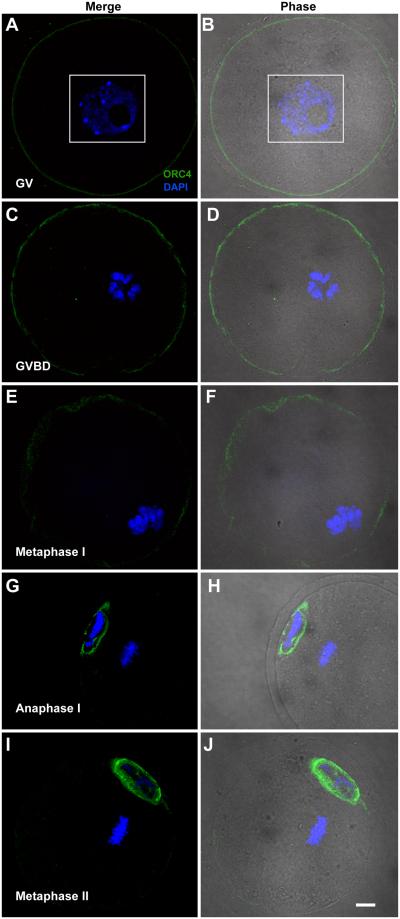
The results described above suggested that ORC4 may play a role in polar body extrusion. To test this, we examined whether ORC4 localized to the chromatin or perinuclear cytoplasm during the first division of female meiosis. We isolated oocytes at the germinal vesicle (GV) stage which is the point just before metaphase I, and cultured them for 3 to 9 hours until they reached metaphase I and anaphase I. At both the GV and GVBD (germinal vesicle breakdown) stages, ORC4 was present in a thin shell just below the cell membrane (Figs. 4A-D). At metaphase I, this layer became more diffuse (Figs. 4E and F). At anaphase I, ORC4 surrounded the polar body chromosomes, but not the oocyte chromosomes (Figs. 4G and H), just as in anaphase II (Fig. 3G). Because ORC4 seemed to play similar roles in anaphase I and anaphase II, we tested whether the same was true for ORC2. We found that ORC2 localized between the separating chromosomes of anaphase I (Suppl. Fig. 6), just as it does in anaphase II [Ortega et al., 2012]. Once the first polar body was extruded, at metaphase II, ORC4 surrounded the polar body nucleus (Figs. 4I and J). These data support the possibility that ORC4 plays a role in the extrusion of both polar bodies. Because the ring-like structures for the ORC4 complex were seen by confocal microscope sections, we tested whether the three dimensional structure was actually an ovoid sphere that completely enclosed the polar body chromatin. Figure 5 shows serial confocal sections of polar body chromatin in late anaphase I that clearly demonstrate that ORC4 forms a sphere that surrounds one set of chromosomes (Figs. 5C-G) but not the other (Figs. 5A-B).
Figure 4. ORC4 in maturing oocytes.
GV oocytes were isolated and cultured in vitro, then stained for ORC4 and visualized by confocal microscopy. GV (A and B), and GVBD oocytes (C and D) contained ORC4 in the cell periphery. Metaphase I oocytes (E and F) had more diffuse patterns of ORC4 distribution. At anaphase I (G and H), ORC4 surrounded the chromatin nearest the oolemma, and this was maintained in metaphase II (I and J). All images are shown at the same magnification, bar = 10 μm.
Figure 5. ORC4 forms an ovoid sphere around the polar body chromatin at anaphase.
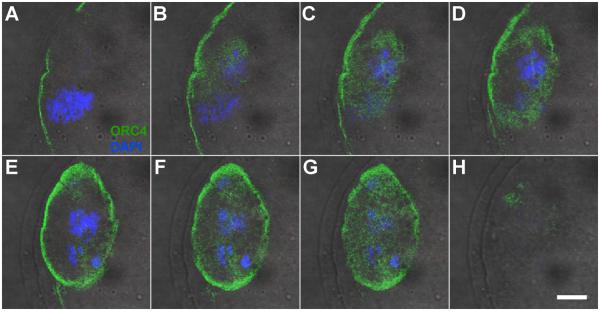
A GV oocyte was matured in vitro until it reached anaphase I and then fixed and stained for ORC4. The images are serial confocal planes starting at one chromosome set at anaphase I (A). The second set of chromosomes appears in (B). ORC4 begins as a small oval shape (B and C) then becomes a ring (D and E) then becomes a solid oval, again (F to G), indicating that ORC4 surrounds only one set of chromosomes as an ovoid sphere at anaphase I. All images are shown at the same magnification, bar = 10 μm.
ORC4 translocates to the chromatin in late G1 to zygotic metaphase
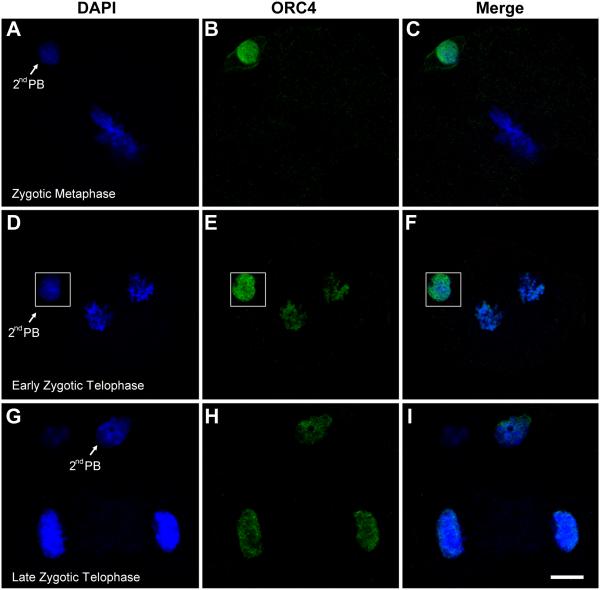
Because ORC4 is a nuclear protein normally associated with DNA replication, it was unexpected to find it primarily in the peri-nuclear cytoplasm during female meiosis and in zygotic G1. We were therefore interested in determining whether it associates with the nuclear DNA at later stages of development. The behavior of ORC4 was different in the 2nd polar body than in the zygotic DNA. ORC4 translocated from the cytoplasm to the nucleus of the 2nd polar body during G1 (Fig. 3L and Q). We never detected ORC4 in the nucleus of the first polar body, and this may be because it degenerated before zygotic metaphase. In some cases when both polar bodies were present at zygotic telophase, we found that ORC4 remained in the cytoplasm of one of them (presumed to be the first polar body) while it had translocated into the nucleus of the other one (presumed to be the second polar body (Suppl. Fig. 7).
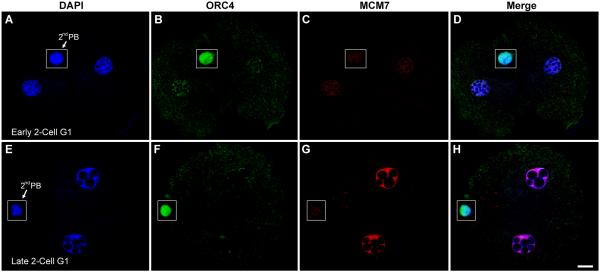
However, we detected no ORC4 associated with the zygotic DNA throughout G1 and metaphase, even though it was clearly detectable in the polar body nucleus (Fig. 6B). By late telophase of the zygote, ORC4 coated all the chromosomes destined for both cells (Fig. 6E) and remained in the newly forming nuclei at late telophase (Fig. 6H). In the 2-cell embryos, ORC4 was localized to the nuclei in early G1, although in much lower levels than was present in the second polar body (Fig. 7B). By late G1, ORC4 had become almost undetectable in the 2-cell embryo nuclei, even though it was still clearly present in the polar body (Fig. 7F). By this time, MCM7 had filled the nuclei (Fig. 7G).
Figure 6. ORC4 translocates to the DNA during zygotic metaphase.
ICSI generated zygotes were fixed and stained for ORC4 at different stages of zygotic metaphase. In zygotic metaphase, ORC4 is present in the nucleus of the second polar body (2nd PB) but not on the zygotic metaphase chromosomes (A-C). By early telophase, both sets of chromosomes are coated with ORC4 (D-F), and this persists to late telophase (G-I). All images are shown at the same magnification, bar = 10 μm.
Figure 7. ORC4 is present in early G1 2-cell embryo nuclei.
ICSI generated 2-cell G1 embryos were fixed and stained for ORC4 and MCM7. In early 2-cell G1, ORC4 is detectable in both nuclei (A-D). Later in G1, MCM-7 is visible in both pronuclei, but ORC4 is absent (E-H). The polar body (PB) retained ORC4, and did not accumulate MCM7. Insets show areas from different confocal planes than the rest of the figure. All images are shown at the same magnification, bar = 10 μm.
Discussion
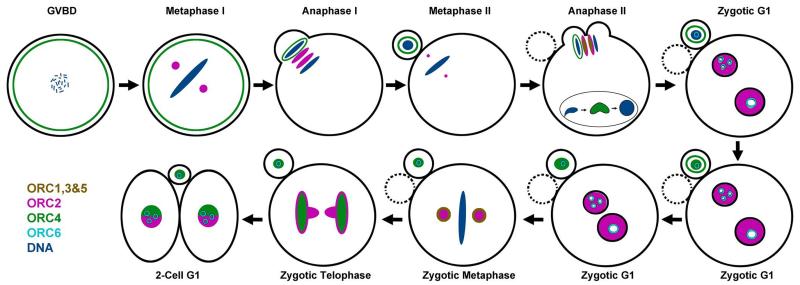
This work represents the completion of our study on the localization of the six ORC proteins during the first cell cycle of the mouse zygote. The data from this and our previous study [Ortega et al., 2012] are summarized in Fig. 8.
Figure 8. Diagram of ORC subunit distribution during the first two cell cycles of the mouse embryo.
The distribution of the ORC subunits were determined in both this study and our previous publication for ORC2 [Ortega et al., 2012].
ORC1-3, 5 and 6
In the zygote, perhaps more than in other cell types, ORC proteins seem to play roles in addition to their well documented function in DNA replication. ORC1-3 and 5 all localize to the area between the separating chromosomes in anaphase II just after fertilization, and to the spindle poles in zygotic metaphase. It is not clear what role these four ORC proteins play in these structures, but it is intriguing that all four localize at both. A recent study has suggested that ORC2-5 assemble in the cytoplasm before being translocated to the nucleus [Ghosh et al., 2011]. The co-localization of ORC1-3 and 5 at the mitotic spindle and between the separating chromosomes at anaphase II suggest that these are storage sites for the initial ORC complexes in close proximity to the newly forming nucleus. A similar localization of ORC2 between separating chromosomes at mitosis was previously noted in HeLa cells [Prasanth et al., 2004]. Interestingly, ORC2 was also found at the spindle poles in the oocyte at metaphase I and between the separating chromosomes of anaphase I (Suppl. Fig. 6). Because both structures disappear and then re-establish in the second mitotic division of maternal meiosis, this suggests that ORC2 plays a role in mitosis other than licensing [Pflumm and Botchan, 2001]. We also demonstrated that ORC1 surrounded the separating chromosomes in zygotic metaphase (Suppl. Fig. 3) just as we had previously shown for ORC2 [Ortega et al., 2012]. ORC2 was constantly found around the DNA during telophase before it moved into the nucleus, and ORC1 seems to follow this pattern. We found that ORC6 was localized to the nucleoli of both pronuclei and the polar body, which agrees with studies in Drosophila [Balasov et al., 2009] and human cells [Thomae et al., 2008].
We were not able to visualize ORC1-3 or 5 co-localized with the DNA at any time. However, recent models for ORC suggest it might be more transiently associated with DNA than previously thought [Li and Stillman, 2012], so it is possible that we missed time points in which ORC subunits were associated with DNA. Our own data for ORC4 (discussed below) and ORC2 (discussed in ref. [Ortega et al., 2012]) suggest that this is particularly true for the zygotes. Additionally, the fully formed ORC is a circular complex in which the six subunits are closely associated [Chen et al., 2008]. This may provide steric hindrance from the other ORC subunits that prevents the antibodies from finding their epitopes.
ORC4 and the Polar Body
The most unexpected finding in our study was the association between ORC4 and the polar body chromatin. We have shown that ORC4 is part of a structure that surrounds the chromatin as an ovoid sphere that is destined to become the first and second polar bodies in both female meiotic divisions. It does not appear that any of the other ORC subunits are involved in this. ORC2 is located adjacent to the ORC4 sphere between the separating chromosomes in both divisions, and ORC1, ORC3, and ORC5 are similarly located there in anaphase II. This is consistent with the recent demonstration that in the first step of the formation of the ORC is the formation of the ORC2-5 complex, to which ORC4 binds before being transported into the nucleus [Ghosh et al., 2011]. This suggests that ORC4 is capable of existing in the cytoplasm separately from the other ORC subunits as observed during polar body formation where an ORC4 sphere was present as soon as the chromosomes began to separate. In the first meiotic division, ORC4 was clearly present in a thin layer just below the oolemma then moved to the separating chromosomes in anaphase. In metaphase II, ORC4 was visible only in the cytoplasm of the first polar body, where it remained during anaphase II in those cases where the first polar body survived. These results raise the intriguing question that cytoplasmic ORC4 plays a role in the separation of the polar bodies from the oocyte.
Polar body extrusion is tightly coupled with the movement of the mitotic plate close to the oolemma during metaphase I and metaphase II in the maturing oocyte [Maro and Verlhac, 2002]. A cortical domain forms over the spindle near the oolemma, and the mitotic plate is oriented perpendicular to the membrane [Longo and Chen, 1985; Maro et al., 1986]. Formin-2, a microfilament binding protein, directs the migration of the mitotic plate to the oolemma [Leader et al., 2002]. A myosin ring helps to define the polar body cytoplasm, and the final emission [Deng et al., 2007]. From these studies, it would appear that the chromatin that is destined to be extruded in the polar body is defined not by a molecular signal but by the orientation of the mitotic plate in the cytoplasm: the chromatin that is nearest to the oolemma will be extruded. The role that ORC4 may play in this, therefore, is not readily apparent. One possibility is that the ORC4 sphere stabilizes the polar body cytoplasm that will form the polar body. In support of this, it is interesting to note that the decondensing sperm nucleus also forms a cone, similar to the polar body, that is subsequently not extruded and reabsorbed back into the oocyte [Deng et al., 2007]. It is not clear why the sperm cone is not extruded but the polar body cone is. We only found the ORC4 sphere around the polar body chromatin, and never around the sperm nucleus, and it is possible that the ORC4 in the polar body cytoplasm plays a role in the eventual extrusion of the polar body cone. Another possibility is that it helps to segregate the polar body chromatin from nuclear factors that would activate genes inappropriately. We are currently testing whether ablation of ORC4 will prevent polar body formation.
ORC4 and Zygotic Chromatin
The role that ORC4 plays in polar body emission seems to be separate from its role in DNA replication. We have previously shown that the second polar body occasionally undergoes visible DNA replication [Yamauchi et al., 2007] so it is likely that enough ORC4 binds to the DNA of the polar body to ensure at least partial licensing. We found that ORC4 undergoes two cytoplasmic to nuclear translocations in the first two embryonic cell cycles. We found three examples of ORC4 being associated with the sperm chromatin and both sets of separating maternal chromosomes at anaphase II (Fig. 3 and Suppl. Fig. 5). In all three cases, ORC4 bound to the DNA of the chromatin at the same time that it formed the ORC4 sphere that surrounded the chromosomes destined to become the polar body. Interestingly, at this point in fertilization, the maternal chromosomes are in anaphase and are not contained within a nuclear membrane therefore these chromosome sets are directly associated with the cytoplasm. It should be noted, the decondensing sperm nucleus also does not have a fully formed nuclear envelope [Usui et al., 1997]. This association appeared to be short-lived, consistent with recent studies suggesting that the ORC complex forms transient structures in some cases [Li and Stillman, 2012; Sonneville et al., 2012]. Another possibility, mentioned above, is that ORC4 remains on the chromatin structures but is no longer available for antibody binding in the fully formed ORC. In this latter case, the data would suggest that ORC4 plays a role in establishing the fully formed ORC as observed in S. pombe [Chuang and Kelly, 1999].
The second obvious association of ORC4 with the chromatin was in late zygotic anaphase. The chromosomes became coated with ORC4 as they were separating from the mitotic plate (Fig. 6). This was preceded by a clear translocation of the cytoplasmic ORC4 in the second polar body into the nucleus just before zygotic metaphase (Figs. 6B and E). Interestingly, in those cases where the first polar body survived unit zygotic metaphase, ORC4 translocated into the nucleus of second polar body, but not the first (Suppl. Fig. 7). In this case, the polar body DNA is sequestered inside nuclear envelope, but the zygotic anaphase chromosomes are not. The data suggest that the roles of ORC4 in the cytoplasm, within the nucleus, and in mitotic chromosomes are regulated both spatially and temporally. It is likely that the association of ORC4 with the chromatin is related to licensing. ORC4 is the only ORC subunit that we have localized to the zygotic anaphase chromosomes. ORC1 (Suppl. Fig. 3) and ORC2 [Ortega et al., 2012] surround the chromosomes but do not appear to bind directly to the DNA at this point. This raises the intriguing possibility that ORC4 directs the formation of ORC on the chromatin. Most current work on human ORC suggests that it is ORC1 that directs the binding of the final complex to DNA. However, in the yeast S. pombe, ORC4 has a unique domain that directs the binding of the ORC to the DNA [Chuang and Kelly, 1999]. It is possible that mouse ORC4 plays a greater role in ORC binding to DNA that human ORC4.
Conclusions
Our data suggest that the ORC subunits play important roles in the mouse zygote other than licensing DNA replication origins. ORC1-3, 4 and 5 may all be involved in the separation of chromosomes during meiosis, and ORC4 may play a role in polar extrusion. Both functions are related to the proper segregation of the chromosomes just before and after fertilization.
Supplementary Material
Acknowledgments
Grant Support:
This work was supported by NIH Grant HD060722 to W.S.W. and by a Vietnamese Education Foundation Fellows grant to Hieu Nguyen
References
- Balasov M, Huijbregts RP, Chesnokov I. Functional analysis of an Orc6 mutant in Drosophila. Proc Natl Acad Sci U S A. 2009;106:10672–7. doi: 10.1073/pnas.0902670106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Speck C, Wendel P, Tang C, Stillman B, Li H. The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2008;105:10326–31. doi: 10.1073/pnas.0803829105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Kelly TJ. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci U S A. 1999;96:2656–61. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Suraneni P, Schultz RM, Li R. The Ran GTPase mediates chromatin signaling to control cortical polarity during polar body extrusion in mouse oocytes. Dev Cell. 2007;12:301–8. doi: 10.1016/j.devcel.2006.11.008. [DOI] [PubMed] [Google Scholar]
- DePamphilis ML. Cell cycle dependent regulation of the origin recognition complex. Cell Cycle. 2005;4:70–9. doi: 10.4161/cc.4.1.1333. [DOI] [PubMed] [Google Scholar]
- Dhar SK, Yoshida K, Machida Y, Khaira P, Chaudhuri B, Wohlschlegel JA, Leffak M, Yates J, Dutta A. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell. 2001;106:287–96. doi: 10.1016/s0092-8674(01)00458-5. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Vassilev AP, Zhang J, Zhao Y, DePamphilis ML. Assembly of the human origin recognition complex occurs through independent nuclear localization of its components. J Biol Chem. 2011;286:23831–41. doi: 10.1074/jbc.M110.215988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Yanagimachi R. Intracytoplasmic sperm injection in the mouse. Biology of Reproduction. 1995;52:709–20. doi: 10.1095/biolreprod52.4.709. [DOI] [PubMed] [Google Scholar]
- Kneissl M, Putter V, Szalay AA, Grummt F. Interaction and assembly of murine pre-replicative complex proteins in yeast and mouse cells. J Mol Biol. 2003;327:111–28. doi: 10.1016/s0022-2836(03)00079-2. [DOI] [PubMed] [Google Scholar]
- Krude T. Initiation of chromosomal DNA replication in mammalian cell-free systems. Cell Cycle. 2006;5:2115–22. doi: 10.4161/cc.5.18.3248. [DOI] [PubMed] [Google Scholar]
- Leader B, Lim H, Carabatsos MJ, Harrington A, Ecsedy J, Pellman D, Maas R, Leder P. Formin-2, polyploidy, hypofertility and positioning of the meiotic spindle in mouse oocytes. Nat Cell Biol. 2002;4:921–8. doi: 10.1038/ncb880. [DOI] [PubMed] [Google Scholar]
- Li H, Stillman B. The origin recognition complex: a biochemical and structural view. Subcell Biochem. 2012;62:37–58. doi: 10.1007/978-94-007-4572-8_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo FJ, Chen DY. Development of cortical polarity in mouse eggs: involvement of the meiotic apparatus. Dev Biol. 1985;107:382–94. doi: 10.1016/0012-1606(85)90320-3. [DOI] [PubMed] [Google Scholar]
- Maro B, Johnson MH, Webb M, Flach G. Mechanism of polar body formation in the mouse oocyte: an interaction between the chromosomes, the cytoskeleton and the plasma membrane. J Embryol Exp Morphol. 1986;92:11–32. [PubMed] [Google Scholar]
- Maro B, Verlhac MH. Polar body formation: new rules for asymmetric divisions. Nat Cell Biol. 2002;4:E281–3. doi: 10.1038/ncb1202-e281. [DOI] [PubMed] [Google Scholar]
- Nordman J, Orr-Weaver TL. Regulation of DNA replication during development. Development. 2012;139:455–64. doi: 10.1242/dev.061838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega MA, Marh J, Alarcon VB, Ward WS. Unique Pattern of ORC2 and MCM7 Localization During DNA Replication Licensing in the Mouse Zygote. Biol Reprod. 2012;87:1–9. doi: 10.1095/biolreprod.112.101774. 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflumm MF, Botchan MR. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development. 2001;128:1697–707. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004;23:2651–63. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Gilbert DM. The many faces of the origin recognition complex. Curr Opin Cell Biol. 2007;19:337–43. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Scholefield G, Veening JW, Murray H. DnaA and ORC: more than DNA replication initiators. Trends Cell Biol. 2011;21:188–94. doi: 10.1016/j.tcb.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Sonneville R, Querenet M, Craig A, Gartner A, Blow JJ. The dynamics of replication licensing in live Caenorhabditis elegans embryos. J Cell Biol. 2012;196:233–46. doi: 10.1083/jcb.201110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer J, Kneissl M, Putter V, Grummt F. Identification and characterization of MmORC4 and MmORC5, two subunits of the mouse origin of replication recognition complex. Chromosoma. 1999a;108:243–9. doi: 10.1007/s004120050374. [DOI] [PubMed] [Google Scholar]
- Springer J, Nanda I, Hoehn K, Schmid M, Grummt F. Identification and chromosomal localization of murine ORC3, a new member of the mouse origin recognition complex. Cytogenet Cell Genet. 1999b;87:245–51. doi: 10.1159/000015435. [DOI] [PubMed] [Google Scholar]
- Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–43. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- Thomae AW, Pich D, Brocher J, Spindler MP, Berens C, Hock R, Hammerschmidt W, Schepers A. Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc Natl Acad Sci U S A. 2008;105:1692–7. doi: 10.1073/pnas.0707260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui N, Ogura A, Kimura Y, Yanagimachi R. Sperm nuclear envelope: breakdown of intrinsic envelope and de novo formation in hamster oocytes or eggs. Zygote. 1997;5:35–46. doi: 10.1017/s0967199400003543. [DOI] [PubMed] [Google Scholar]
- Vashee S, Simancek P, Challberg MD, Kelly TJ. Assembly of the human origin recognition complex. J Biol Chem. 2001;276:26666–73. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Shaman JA, Boaz SM, Ward WS. Paternal Pronuclear DNA Degradation Is Functionally Linked to DNA Replication in Mouse Oocytes. Biol Reprod. 2007;77:407–415. doi: 10.1095/biolreprod.107.061473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.