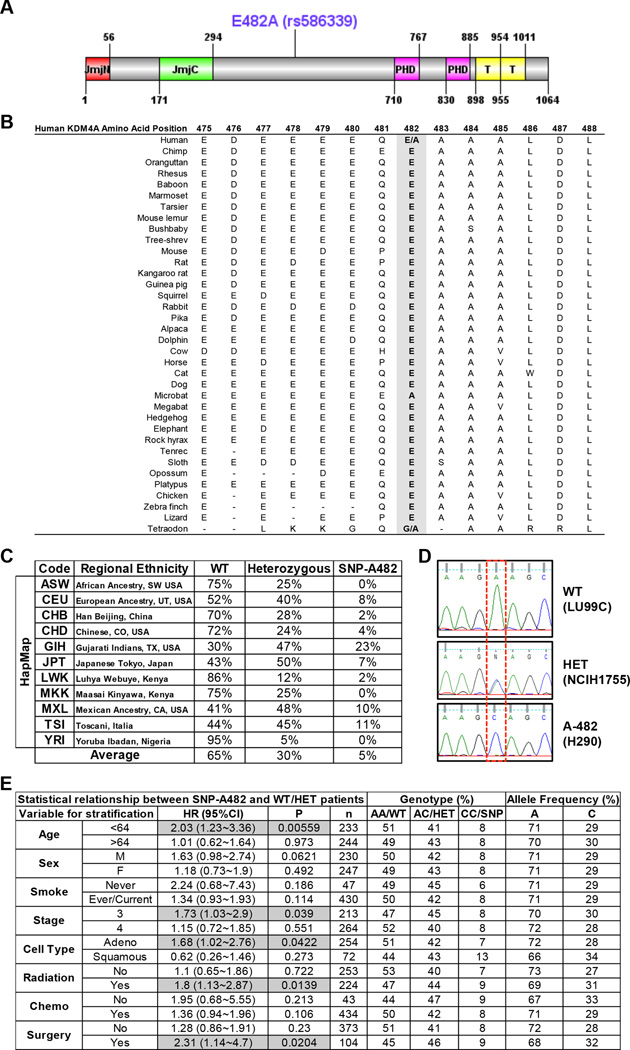
Figure 1. KDM4A SNP-A482 (rs586339) correlates with worse outcome in NSCLC patients.
(A) Schematic of the human KDM4A protein is shown with both the protein domains and the position of the coding SNP rs586339 (E482A). Jumonji (JmjN and JmjC), PHD and Tudor (T) domains are represented. (B) E482 is the conserved allele. The alignment of sequence surrounding E482A is shown for multiple species. (C) HapMap frequencies for rs586339 are presented (August 2010 HapMap public release #28) (13). ASW- African Ancestry in SW USA (n=57); CEU- U.S. Utah residents with ancestry from northern and western Europe (n=113); CHB- Han Chinese in Beijing, China (n=135); CHD- Chinese in Metropolitan Denver, CO, USA (n=109); GIH- Gujarati Indians in Houston, TX, USA (n=99); JPT- Japanese in Tokyo, Japan (n=113); LWK- Luhya in Webuye, Kenya (n=110); MKK- Maasai in Kinyawa, Kenya (n=155); MXL- Mexican Ancestry in Los Angeles, CA, USA (n=58); TSI- Toscani in Italia (n=102); YRI- Yoruba in Ibadan, Nigeria (n=147). (D) Representative KDM4A sequencing plots from three different lung cancer cell lines- homozygote wild type (WT, GAA:GAA), heterozygote (HET, GAA:GCA) and homozygote SNP (A-482, GCA:GCA). (E) KDM4A SNP-A482 stratification for late stage NSCLC patients. HR represents the hazard ratio, 95%CI the 95% confidence interval, p the p value, n the number of patients in each category, AA the genotype for homozygote wild-type (E482), AC for heterozygote, CC for homozygote SNP (A482). Gray boxes highlight the parameters with significance, including the data in panels B-F. Frequency comparisons were tested using the chi-square test, and Hazard Ratios were calculated using a Cox model. Also see supplementary Figure S1.