Figure 1.
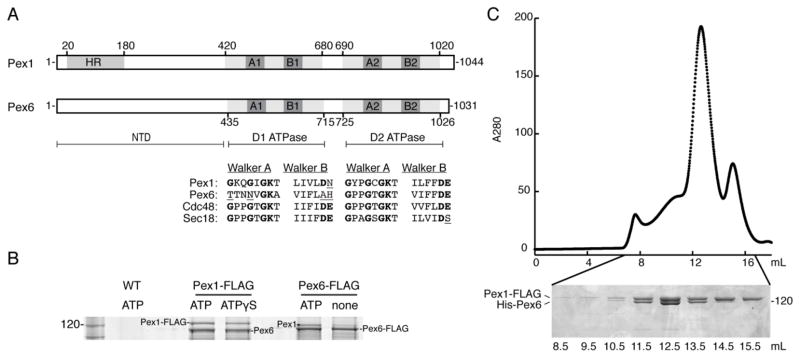
A) Schematic representation of Pex1 and Pex6 from S. cerevisiae. Both are Type-2 AAA+ ATPases with an N-terminal domain (NTD) and two ATPase domains, D1 and D2. HR: N-terminal region homologous to the N-domain of p97 and NSF. A1 and A2: Walker A motif (GxxGxGKT) in the D1 or D2 ATPase domain. B1 and B2: Walker B motif (ϕϕϕϕDE, ϕ: hydrophobic amino acid) in the D1 or D2 ATPase domain. The alignment shows the conservation of the Walker A and Walker B motifs in ScPex1, ScPex6, ScCdc48, and ScSec18. Poorly conserved residues are underlined.
B) Endogenous Pex1 and Pex6 from S. cerevisiae depend on the presence of nucleotide to form a complex. Pex6 co-immunoprecipitated with FLAG-tagged Pex1 in the presence of ATP or ATPγS. Pex1 co-immunoprecipitated with FLAG-tagged Pex6 in the presence of ATP, but this association is diminished when no nucleotide is present. A mock co-immunoprecipitation using the parent wild-type strain with untagged Pex1 and Pex6 served as a control.
C) Recombinant Pex1-FLAG and His-Pex6 purified as a stoichiometric complex from E. coli. Pex1-FLAG and His-Pex6 were co-expressed in BL21* E. coli and subsequently purified by Ni-NTA agarose, anti-FLAG affinity resin, and size exclusion chromatography. The Superose 6 elution profile and SDS-PAGE analysis of the fractions show that the main peak contains both Pex1-FLAG and His-Pex6, and a minor peak represents a smaller homo-oligomer of Pex1-FLAG.
