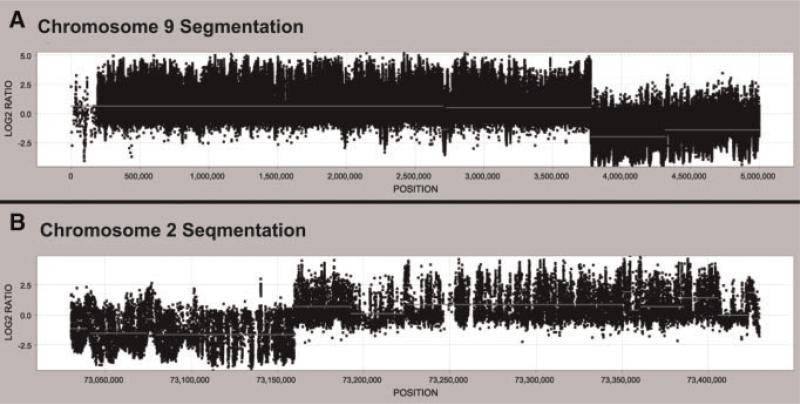
Fig. 2.
Oligonucleotide fine tiling array. A custom NimbleGen fine tiling array spanning positions chr2: 73,030,173–73,429,557 and chr9: 1–5,000,000 was used to map the rearrangement breakpoints. The Cy3/Cy5 log 2 ratios [Chr.9 Der./(Chr2 + Chr9)] deviate significantly from 0 at chr2: 73,159,900 and ~chr9: 3,775,600, indicating the approximate rearrangement breakpoints.