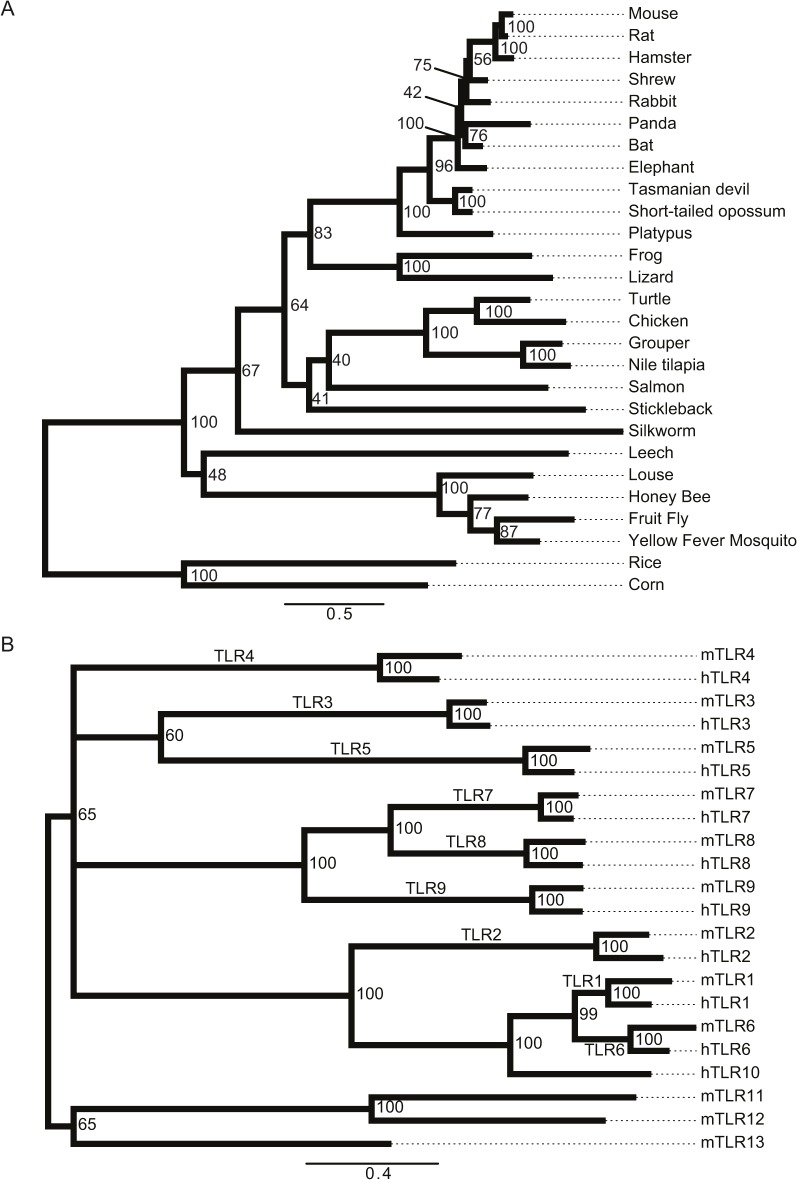
Fig 7. Phylogenetic analysis of TLR13 in species and of the TLR repertoires in humans and mice.
(A) The phylogenetic tree displays a choice of species where we identified an ortholog to TLR13 using the OMA browser and NCBI Blast search. The hits were confirmed with FACT to test whether the predicted orthologs have similar feature architecture as the query protein. Thus, the orthologs are firstly similar in sequence and secondly similar in their feature architecture. Numbers at the branch points indicate bootstrap values. (B) The phylogenetic tree displays evolutionary relationship of human and mouse TLR proteins. The tree was constructed as described in (A). Note that if a particular TLR is found in both species then both orthologs exhibit very close evolutionary relationship (bootstrap value 100). The nucleic acid-sensing TLRs do not cluster in one branch: TLR7, TLR8 and TLR9 are found in a different branch than TLR3, and the mouse-specific TLR13 clusters yet in another branch together non-nucleic acid sensors TLR11 and TLR12. Numbers at the branch points indicate bootstrap values (cut-off > 50).