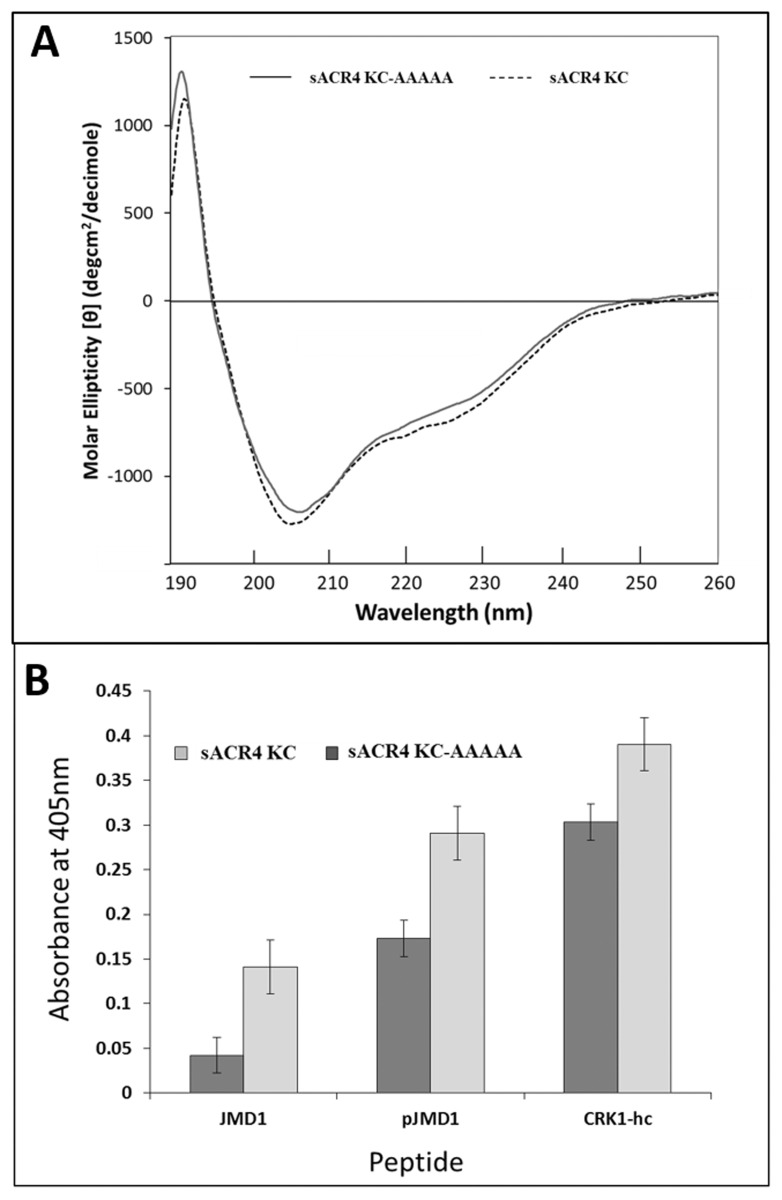
Fig 6. Binding of ‘KDSAF’ peptides to sACR4 KC and sACR4 KC-AAAAA.
(A) Conformational analysis of sACR4 KC 9 (dashed line) and sACR4 KC-AAAAA (solid line) indicating that mutations did not cause protein misfolding. Far UV CD spectra in 10 mM Tris (pH 7.4) and 0.1 mM TCEP with an A280 value of ~1.0. (B) Peptide binding assay demonstrating the binding efficiency of ‘KDSAF’ containing peptides to the sACR4 KC protein and sACR4 KC-AAAAA immobilized on a Ni2+ coated plate. The sACR4 KC-AAAAA mutant shows reduced binding affinities for ‘KDSAF’ containing peptides when compared to wild-type sACR4. Each peptide binding experiment was performed in triplicate and data represented as the mean ± the S.E. Data were corrected for dilution factor and subtracting the absorbance of the control (SUMO) from the experimental samples.