Extended Data Figure 6. Molecular features of HCS m6A-switches.
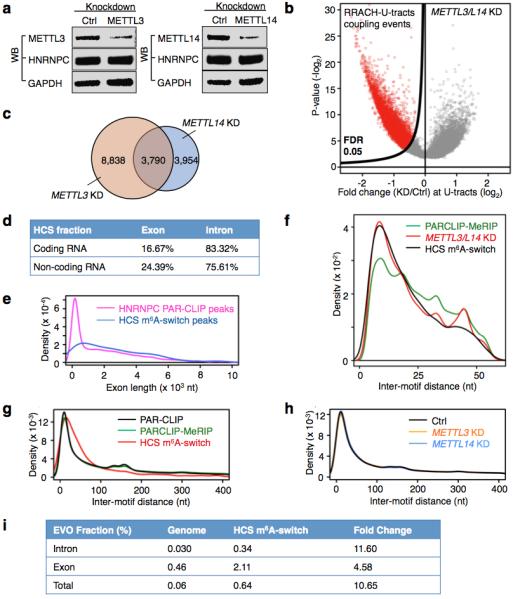
a, Western blot showing stable hnRNP C protein abundance upon METTL3/L14 KD. b, Volcano plot of the METTL3/L14 KD data depicting RRACH-U-tracts coupling events (unfilled red circles) as defined in Extended Data Fig. 4b, according to their p-values39 (P, y-axis) and fold change values at the U-tracts (E, x-axis). c, Overlap of RRACH-U-tract coupling events with decreased hnRNP C binding by METTL3 and METTL14 KD. d, The intron fraction of HCS m6A-switches in coding RNA and non-coding RNA. e, Density plot displaying the distribution of exonic m6A-switches/HNRNPC PAR-CLIP peaks according to exon length. f, Inter-motif (RRACH-U-tract) distance distributions suggest that m6A-switches have a preference for shorter distances between the RRACH and U-tract (> 5xU) motifs. The distribution curves are from PARCLIP-MeRIP data (green), METTL3/L14 KD (red) and HCS m6A-switches (black). g, Analysis of the inter-motif (U-tract-U-tract) distance patterns, previously identified by iCLIP20, in PARCLIP-MeRIP, METTL3/L14 KD and HCS m6A-switch data. The peaks at ~165 and ~300 nucleotides are clearly present. For the 2,798 high confident switches, we analyzed those in which the other U-tract motif is also in a PAR-CLIP-identified sequence; the long-range peaks seem to have shifted to longer distances (~220 and ~370 nucleotides). h, METTL3/L14 KD does not affect the inter-motif (U-tract-U-tract) distance distributions for U-tracts (≥5x U) in HEK293T cells. i, EVOfold analysis for the 2,798 HCS m6A-switches. The chances for HCS m6A-switches to have EVOfold records are significantly higher than random genomic sequences. We first calculated the number of HCS sites in the EVO database if occurring in random to be ~1.7 HCS sites. We found 18 HCS sites are present in EVO database, resulting in ~11x enrichment. This result is further divided into intronic and exonic regions.
