Extended Data Figure 7. m6A-switches regulate the abundance of target mRNAs.
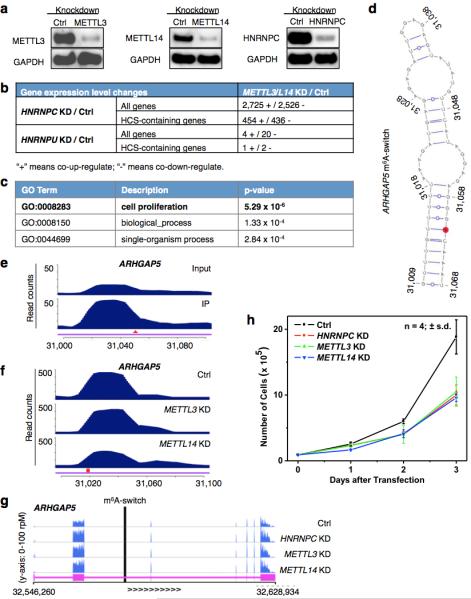
a, HNRNPC, METTL3/L14 KD confirmed by Western blots. b, HNRNPC KD and METTL3/L14 KD co-regulated the expression of a large number of genes. Gene expression changes between Ctrl and HNRNPC, HNRNPU, METTL3/L14 KD HEK293T cells were analyzed by Cuffdiff239,40, and the absolute numbers of differential expressed genes are shown. HCS-containing genes refer to the 1815 genes containing high confident m6A-switches. The RNA-seq data from HNRNPU KD HEK293T cells (GEO34995 dataset41) was analyzed for comparison with a different mRNA binding protein. HnRNPU did not show preferential interaction with the 2,577-m6A modified MALAT1 hairpin (Fig. 1b, 1c). c, Gene ontology analysis of the m6A-switch-containing genes whose expression level were co-differentially-regulated by HNRNPC and METTL3/L14 KD, against all m6A-switch-containing genes as background. d, An example of m6A-switch among co-regulated transcript is the ARHGAP5 transcript (NM_001030055). Its proposed secondary structure with m6A methylation site in red is shown with the opposing the U-tract in a stem. e-f, PARCLIP-MeRIP detected positive IP/Input enrichment at the RRACH site (red arrowhead) of the ARHGAP5 m6A-switch (e), while METTL3/L14 KD decreased hnRNP C binding at the U-tract (red square) of this m6A-switch (f). g, The expression level of ARHGAP5 gene was co-upregulated by HNRNPC, METTL3/L14 KD, as shown by the RNA-seq data from HEK293T cells. The vertical black line represents the m6A-switch site. h, HNRNPC, METTL3/L14 KD decreased the proliferation rates of HEK293T cells to a similar extent. n = 4, ± s.d., biological replicates.
