Extended Data Figure 8. m6A-switches regulate alternative splicing of target mRNAs.
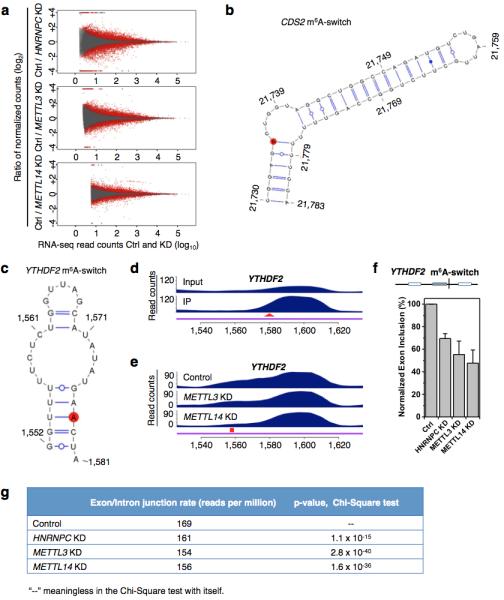
a, Fold changes (KD/Ctrl, log2) in normalized exon expression against RNA-seq reads detect the exons in HNRNPC KD, METTL3 KD, METTL14 KD and control samples. Statistically Significant Differentially Expressed Exons (SSDEE) called by DEXSeq are indicated in red. b, Proposed secondary structure of the CDS2 hairpin with the m6A methylation site shown in red, opposing the U-tract region. Nucleotide position numbers correspond to their locations along the human CDS2 transcript (NM_003818). c, Proposed secondary structure of the YTHDF2 hairpin with the m6A methylation site shown in red, opposing the U-tract region. Nucleotide position numbers correspond to their locations along the human YTHDF2 transcript (NM_001173128). de, PARCLIP-MeRIP detected a positive enrichment at the RRACH site (red arrowhead) (d), while METTL3/L14 KD decreased hnRNP C binding at the U-tract (red square) of this YTHDF2 m6A-switch (e). f, The inclusion level of one YTHDF2 exon is co-down-regulated by HNRNPC KD, METTL3 KD and METTL14 KD, as validated by RT-PCR. n = 3, ± s.d., biological replicates. g, We analyzed our polyA+ RNA-seq data to look for reads that span intron/exon junctions on CDS m6A-switch containing genes. We find that the control sample has significantly higher reads spanning intron/exon junctions than HNRNPC, METTL3/L14 KD samples. This result indicates that m6A depletion at the CDS m6A-switches promotes intron exclusion.
