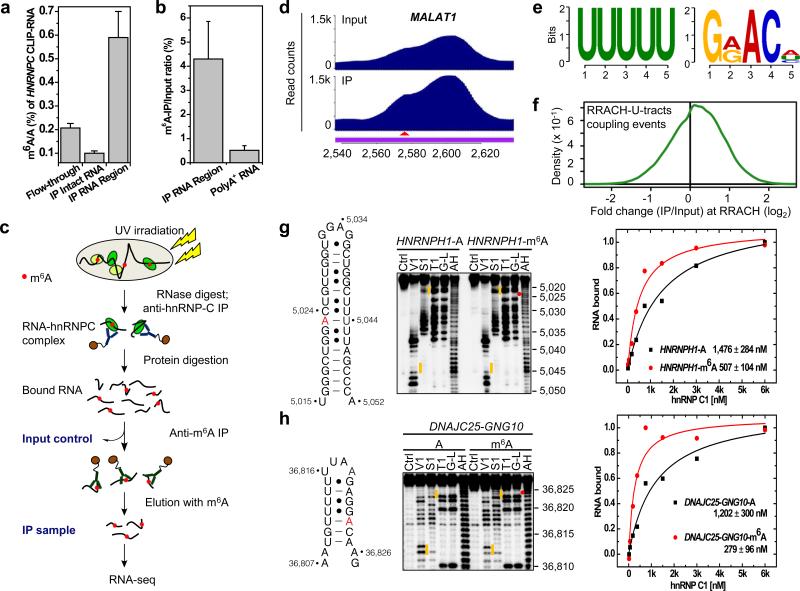
Figure 2. PARCLIP-MeRIP identifies m6A-switches transcriptome-wide.
a, CLIP-2dTLC showing the m6A enrichment in hnRNP C bound RNA regions. n = 3, ± s.d., biological replicates. b, hnRNP C bound RNA regions had higher anti-m6A pull down yield than polyA+ RNA. n = 3, ± s.d., biological replicates. c, Illustration of the PARCLIP-MeRIP protocol. IP, immunoprecipitation. d, PARCLIP-MeRIP identified m6A residue around the MALAT1 2,577 site. e, Binding motifs identified by FIRE with PARCLIP-MeRIP peaks. f, Density plot showing the positive enrichment at RRACH sites. g-h, Validation of two m6A-switches by S1/V1 structural probing and filter binding. n = 4, ± s.d., technical replicates. Same annotation as in Fig. 1c, d.