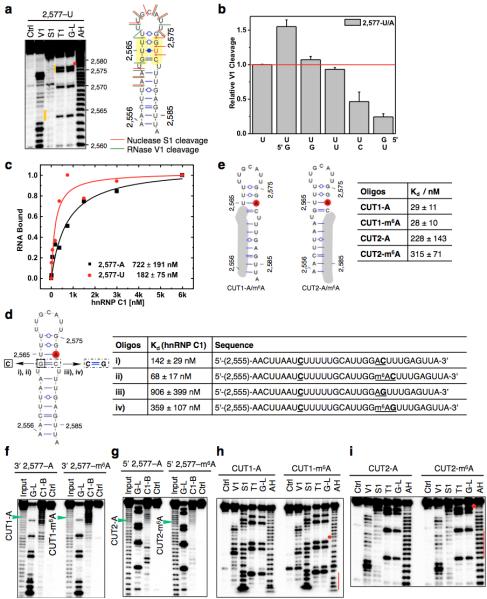
Extended Data Figure 2. Increased accessibility of U-tracts enhances hnRNP C binding.
a, Structure probing of the 2,577A-to-U mutated MALAT1 hairpin (2,577-U), same annotation as in Fig. 1d. b, Quantification of the RNase V1 cleavage signal for the U-tract region from RNA structural mapping assays as in a. To correct for sample loading difference, each band signal was normalized to the band signal of the 3’ most U of the U-tract. n = 2, technical replicates. c, Filter-binding curves displaying the binding affinities between recombinant hnRNP C1 and 2,577-U/A oligos. n = 3, ± s.d., technical replicates. d, Filter-binding results showing the binding affinities between recombinant hnRNP C1 and four mutated MALAT1 oligos. (i) Mutate G-C to C-C, A2,577: predicted to weaken the hairpin stem and increase hnRNP C binding. Results: binding improved from 722 nM Kd to 142 nM (5-fold); (ii) Mutate G-C to C-C, m6A2,577: in this context of weaker stem, m6A is predicted to confer a smaller effect compared to wild-type hairpin. Result: improved binding only 2-fold instead of 8-fold; (iii) Restore C-C to C-G, A2,577: predicted to restore the hairpin stem and decrease hnRNP C binding compared to C-C mutant. Result: binding decreased by 6.4-fold; (iv) Restore C-C to C-G, m6A2,577: in this context of restored stem, m6A is again predicted to confer increased binding compared to A2,577 hairpin. Result: improved binding by 2.5-fold. n = 3 each, ± s.d., technical replicates. e, RNA alkaline hydrolysis terminal truncation assay showing recombinant hnRNP C1 binding to terminal truncated MALAT1 hairpin oligos (2,577 site m6A methylated or unmethylated). In this assay, 3′ radiolabeled MALAT1 2,577 hairpin oligos were terminal truncated by alkaline hydrolysis into RNA fragments which were then incubated with hnRNP C1 protein followed by filter binding wash steps. The remaining RNA on the filter paper was isolated and analyzed by denaturing gel electrophoresis, as indicated in the lane “C1-bound or C1-B”. “Input” refers to alkaline hydrolysis truncated RNA oligos used for incubation with hnRNP C1; “G-L or G-ladder” was generated from RNase T1 digestion; “Ctrl” refers to the intact MALAT1 hairpin without alkaline hydrolysis truncation. One pair of methylated/unmethylated truncated oligos (CUT1, marked by green arrows) was selected for subsequent biochemical analysis, due to their strong interaction with hnRNP C1. f, RNA terminal truncation assay as in e except 5′ 32P-labeled oligos were used. One pair of methylated/unmethylated truncated oligos (CUT2, marked by green arrows) was selected for subsequent biochemical analysis. g, Structure probing of the CUT1 oligos using RNase V1 and nuclease S1 digestion, same annotation as in Fig. 1e. The red dot marks the m6A site and the red line marks the U-tract region. h, Structure probing of the CUT2 oligos using RNase V1 and nuclease S1 digestion, same annotation as in g. i, Truncated oligos with exposed U-tracts increased hnRNP C binding regardless of m6A. n = 3, ± s.d., technical replicates.