Extended Data Figure 3. m6A is enriched in the vicinity of hnRNP C binding sites.
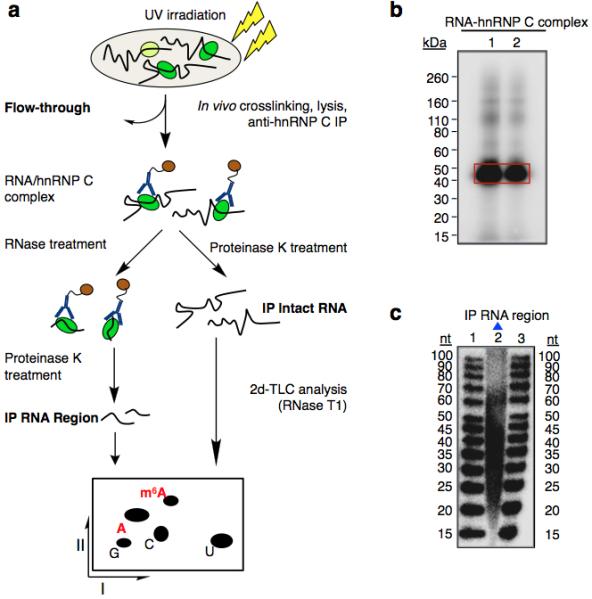
a, Schematic diagram of the CLIP-2dTLC protocol. IP, immunoprecipitation; nt, nucleotide. The RNase T1 used in our 2dTLC assay cleaves single-stranded RNA after guanosines, so the m6A/A ratio determined here represents the m6A fraction of all adenosines following guanosines. b, Analysis of crosslinked RNA-hnRNP C complexes (CLIP RNP) using denaturing gel electrophoresis (lanes 1 and 2). Positions of the protein size standards are shown on the left. hnRNP C IP RNA region (RNA samples within RNA-hnRNP C crosslinked complexes) were extracted from the gel slices marked by the red rectangle. c, Denaturing gel analyzing the size distribution for the hnRNP C PAR-CLIP RNA samples (lane 2). The RNA size standards were loaded in lanes 1 and 3.
