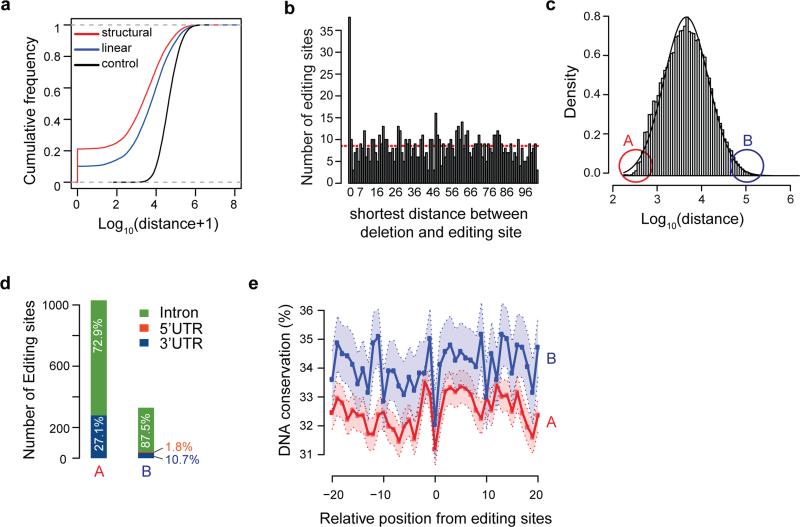
Figure 2. ADAR1 binding signature reflects its function in RNA editing.
(a) Shortest distance between ADAR1-bound Alu sites and RNA editing sites (DARNED database, same below) in the same gene. Linear: linear genomic distance; structural: distance calculated between predicted dsRNA structures harboring the CLIP cluster and editing sites; control: distance between CLIP clusters and random A's chosen from the same region as authentic editing sites. Both linear and structural distances are significantly smaller than control (p < 2.2e-16, Kolmogorov–Smirnov (KS) test). (b) Histogram of distance (up to 100nt) between deletions in CLIP reads and closest RNA editing sites in the same gene. Red dashed line represents the average distance in the range shown. (c) Histogram of closest distance between ADAR1-bound Alu clusters in the same gene. A and B denote the bottom and top 5% of distances respectively. (d) Genomic distribution of editing sites within 100nt of Alu clusters in groups A and B as defined in (c). The distribution of these editing sites in different regions of annotated genes is shown. Note that no editing sites were found in coding exons. (e) Conservation level of positions surrounding editing sites in groups A and B across primates. DNA conservation was calculated as % sequence identify. Light shaded area represents confidence intervals.