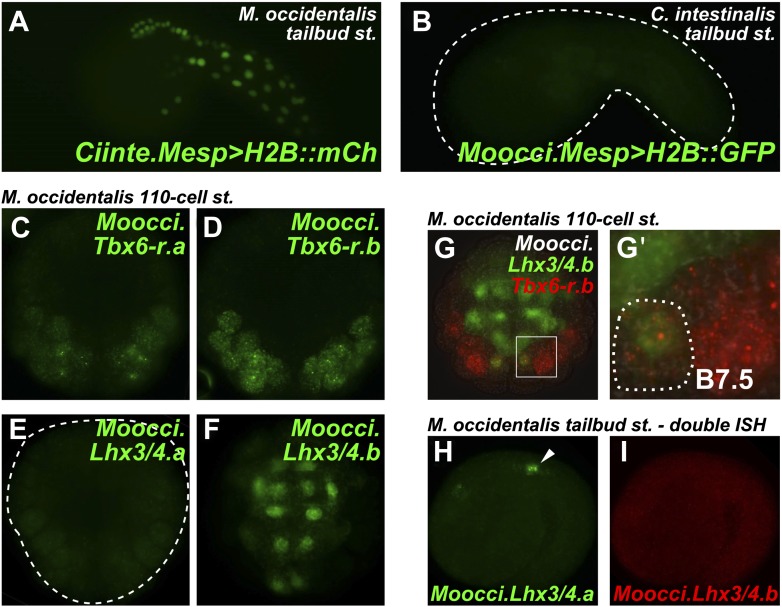
Figure 4. Developmental system drift of Mesp regulation between C. intestinalis and M. occidentalis.
(A) M. occidentalis tailbud embryo electroporated with a Ciinte.Mesp>H2B::mCherry reporter construct. Weak reporter gene expression was observed in the B7.5 lineage and occasionally in other territories including B-line mesenchyme and tail muscle cells, and A-line neural plate derivatives. (B) C. intestinalis tailbud embryo electroporated with Moocci.Mesp>H2B::GFP reporter. No fluorescence was seen in any cells, indicating complete lack of activity of Moocci.Mesp enhancer in wild-type C. intestinalis embryos. (C) In situ hybridization (ISH) for Moocci.Tbx6-r.a, (D) Moocci.Tbx6-r.b, (E) Moocci.Lhx3/4.a, and (F) Moocci.Lhx3/4.b in 110-cell stage embryos. (G) Double ISH in 110-cell stage embryo reveals co-expression of Moocci.Lhx3/4b (green) and Moocci.Tbx6-r.b (red) exactly in the B7.5 cells of M. occidentalis. (G′) Magnified view of inset in (G). (H) Double ISH for Moocci.Lhx3/4.a (green) and (I) Moocci.Lhx3/4.b (red) in a mid-tailbud embryo. Moocci.Lhx3/4.a but not Moocci.Lhx3/4.b is expressed in motor ganglion neurons (arrowhead).
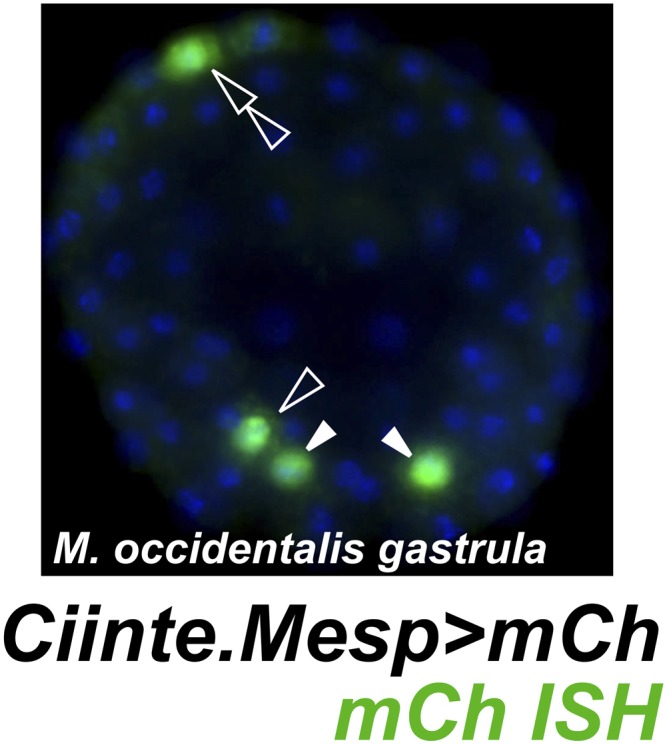
Figure 4—figure supplement 1. Weak and leaky expression of Ciinte.Mesp reporter in M. occidentalis embryos.