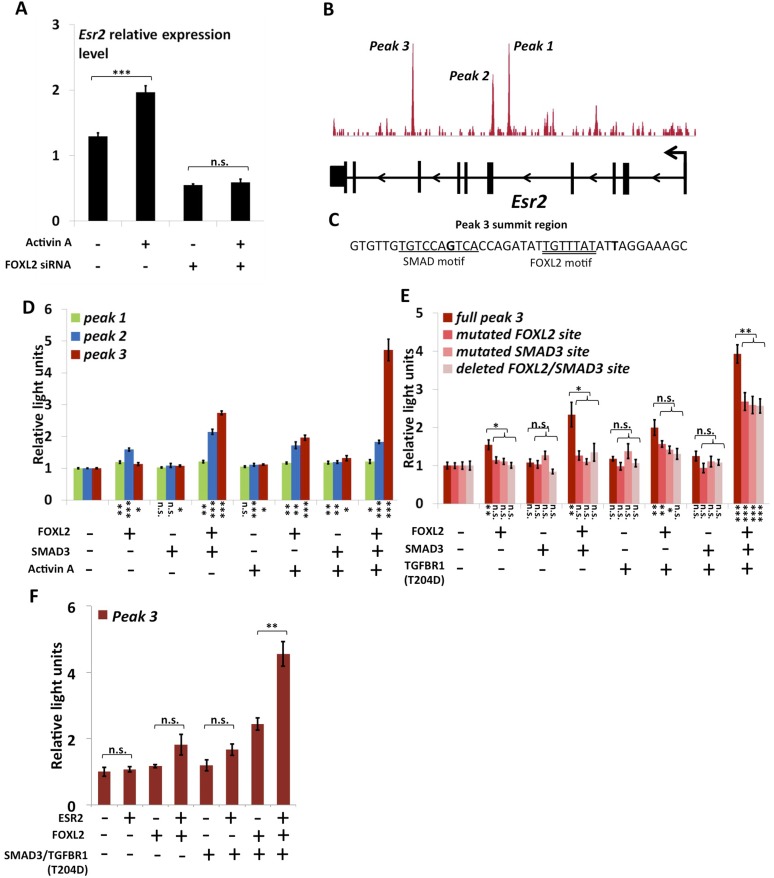
Figure 3. FOXL2 is required for ESR2 expression.
(A) Relative amounts of Esr2 cDNA, determined by qPCR, in mouse follicular cells transfected with an anti-FOXL2 or control siRNAs for 24 hr and treated by 2 nM Activin A or a vehicle for 18 hr. The average expression level of Sdha and Actb was used as a control for normalization. The values are the mean of three independent experiments performed in duplicate. (B) FOXL2 peaks in the Esr2 TU. Three main peaks can be observed in the fourth and eighth introns. (C) Sequence around the summit of the FOXL2 peak in the eighth intron. FOXL2 and SMAD motifs are present near the peak summit. (D–F) Luciferase assays showing FOXL2/SMAD3/ESR2 transcriptional cooperation/synergy on the Esr2 regulatory elements. COV434 cells were transfected with constructs driving the expression of FOXL2, SMAD3, ESR2, and/or a constitutive mutant of TGFBR1 (T204D), as well as with constructs containing the sequences of Peaks 1–3 (D), mutated versions of Peak 3 (E) or Peak 3 alone (F) cloned upstream of a minimal CMV promoter driving the expression of the Firefly luciferase gene, and a construct for the expression of the Renilla luciferase. In the indicated conditions, the cells were treated with 2 nM Activin for 18 hr before lysis. Transcriptional activity was measured as the ratio of Firefly/Renilla luciferases. p-values according to a Student's t test between the indicated conditions and the control condition: *p < 0.05; **p < 0.01; ***p < 0.001; n.s.: non-significant. Asterisks above the histogram in (E) represent the maximum p-value of a Student's t test between full Peak 3 transcriptional activity and all three mutants. Experiments were performed in six replicates.