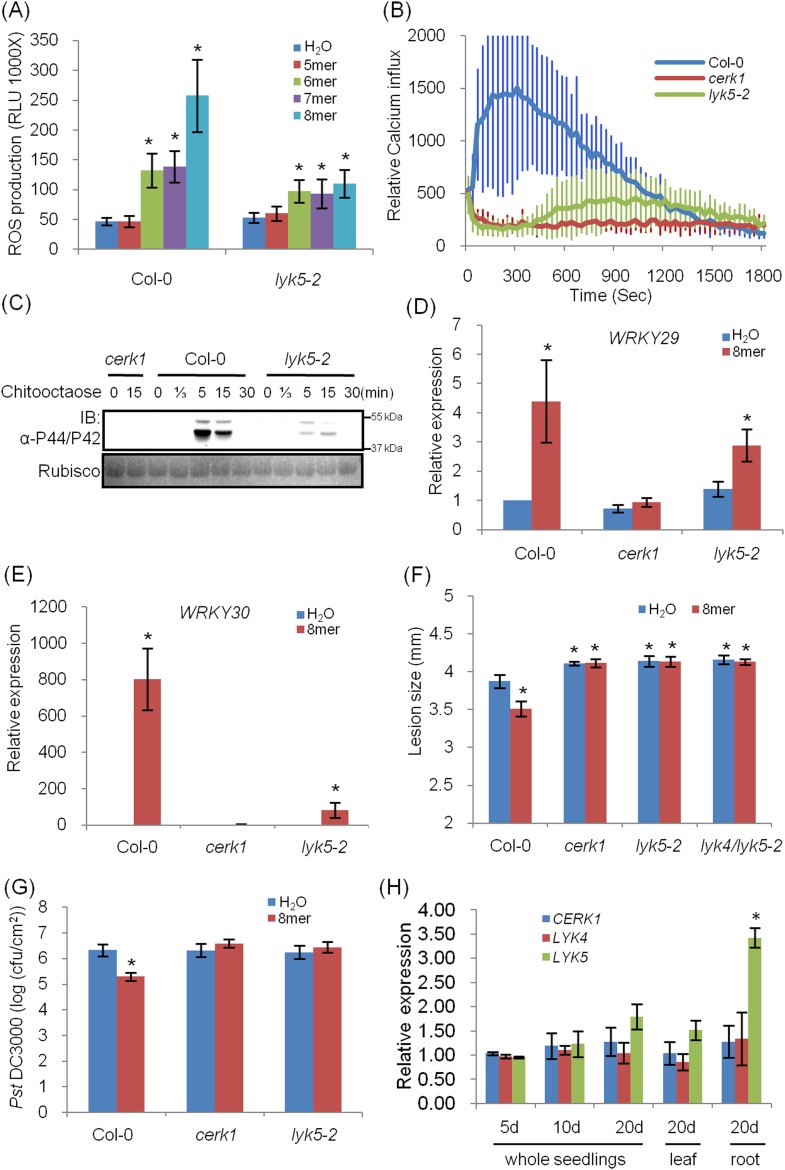
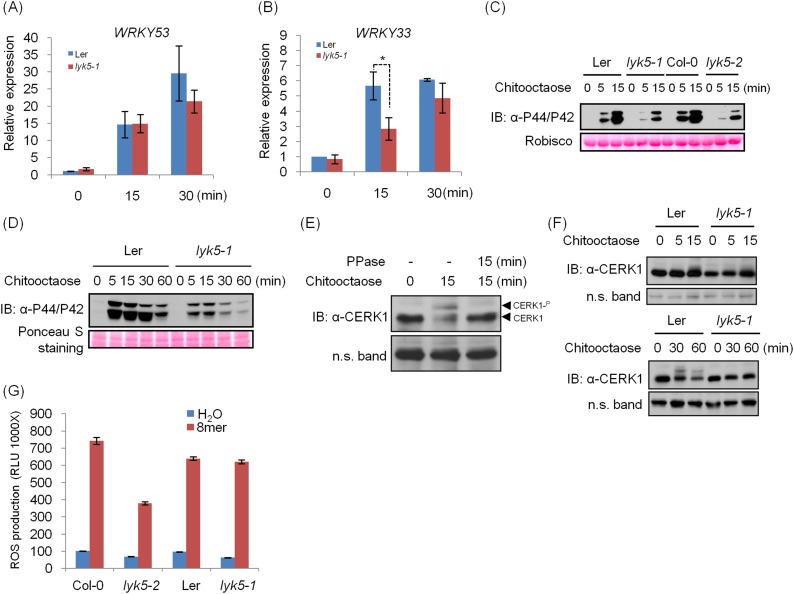
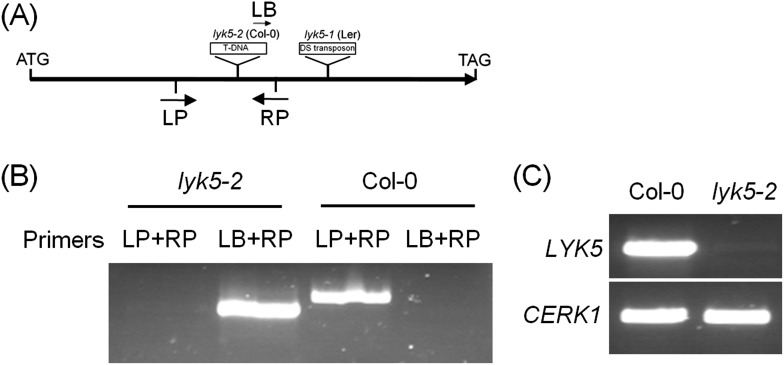
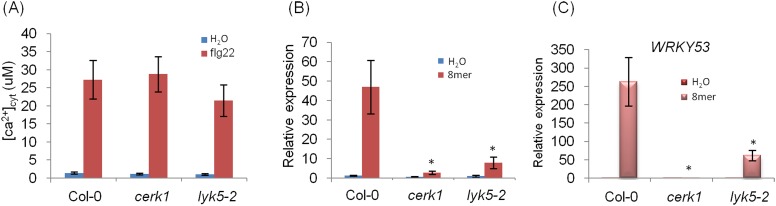
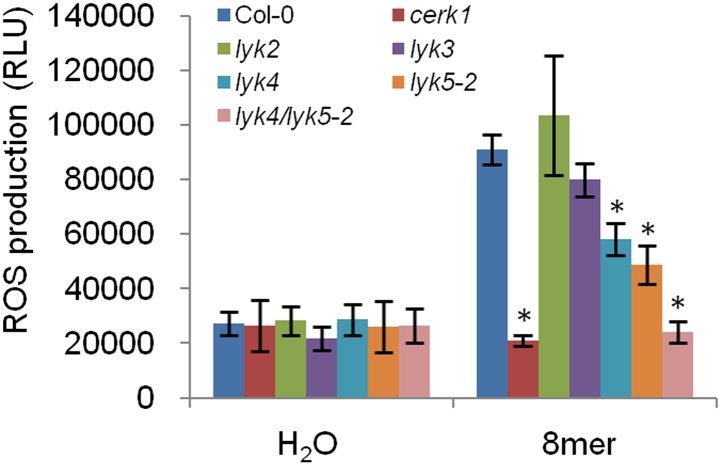
Figure 1. Atlyk5 mutant plants are defective in chitin-triggered immune responses.
(A) ROS production was measured from Col-0 wild-type and Atlyk5-2 mutant plants for 30 min after treatment with different chitin oligomers. 5mer: chitopentaose, 6mer: chitohexaose, 7mer: chitoheptaose, and 8mer: chitooctaose. Data are mean ± SE (n = 8). Asterisks indicate significant difference relative to H2O treated Col-0 wild-type plants. (p < 0.01, Student's t test). (B) Calcium influx in the wild-type, Atcerk1 and Atlyk5-2 mutant plants expressing aequorin was recorded for 30 min after chitooctaose treatment. (C) MAP kinase phosphorylation after chitooctaose treatment was detected by immunoblot using anti-P44/P42 antibody. (D) AtWRKY29 (At4g23550) and (E) AtWRKY30 (At5g24110) gene expression was analyzed using qRT-PCR in the wild-type, Atcerk1 and Atlyk5-2 mutant plants with or without treatment with chitooctaose, 8mer. UBQ10 (At4g05320) was used a control. Data are mean ± SE (n = 3). Asterisks indicate significant difference relative to H2O treated Col-0 wild-type plants. (p < 0.01, Student's t test). (F) 4-week-old leaves from Col-0 wild-type, Atcerk1, Atlyk5-2, and Atlyk4/lyk5-2 mutant plants were inoculated with Alternaria brassicicola 24 hr after hand-infiltration with H2O or 1 µM chitooctaose. The diameter of the lesion area was measured 4 days after inoculation. Data are mean ± SE (n = 12). Asterisks indicate significant difference relative to H2O treated Col-0 wild-type plants. (p < 0.05, Student's t test). (G) Leaf populations of Psuedomonas syringae pv. tomato DC3000 3 days after inoculation. 4-week-old plants were either pretreated with H2O or 1 µM chitooctaose 24 hr before inoculation with P. syringae. Data are mean ± SE (n = 9). Asterisk indicates T-test significant difference compared with H2O-treated Col-0 plants at p < 0.05, Student's t test. (H) AtCERK1, AtLYK4 and AtLYK5 gene expression in different plant ages and plant tissue. RNA from whole seedling of 5 day, 10 day, 20 day old plants and leaf and root tissues from 20 day old plants were used for reverse transcript and qRT-PCR was performed using specific primers. Data are mean ± SE (n = 3). Asterisks indicate significant difference relative to chitiooctaose treated Col-0 wild-type plants (p < 0.01, Student's t test).