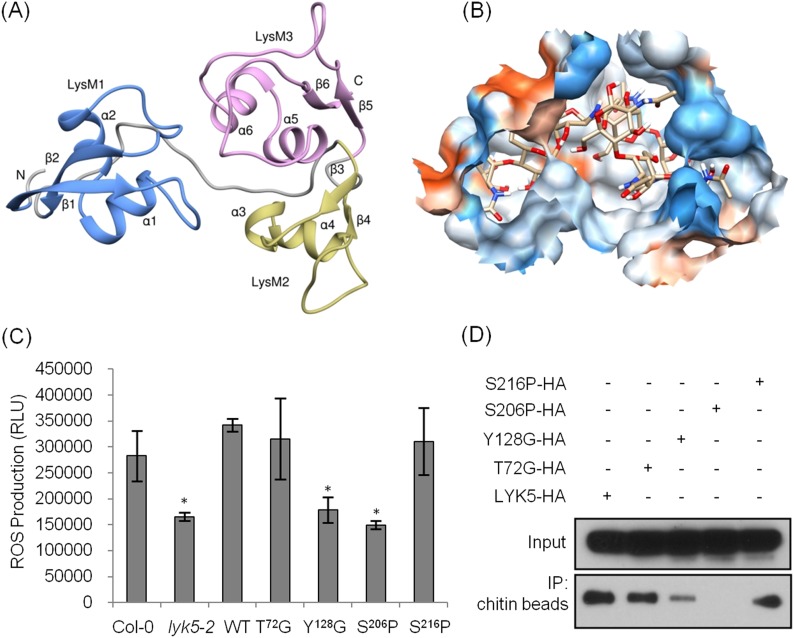
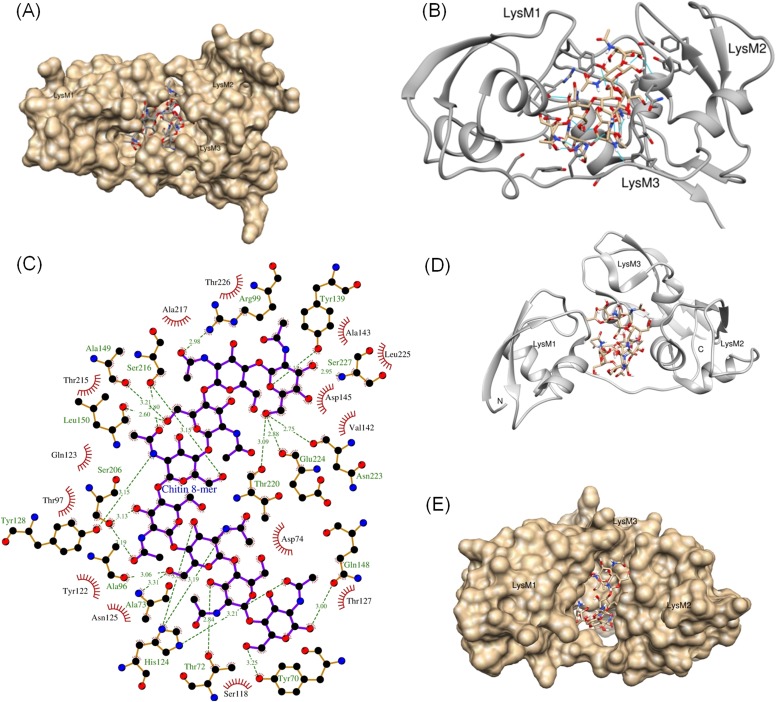
Figure 4. Tyr-128 and Ser-206 are important for AtLYK5-mediated chitin response.
(A) A computational ribbon structure of the AtLYK5 ectodomain was built based on crystal structure of fungal ECP6. The model shows the three AtLYK5 LysM domains, i.e. LysM1-3. Each LysM domain contains two beta strands and two helixes interconnected via loops. (B) The binding affinity was calculated at −8.9 kcal mol−1. The binding site was formed by 3 LysM motifs. Green lines depict hydrogen bonds formed between ligand atoms and their corresponding residues atoms. (C) Reactive oxygen species (ROS) was measured within 30 min after chitin treatment. The AtLYK5 wild-type gene or versions with specific point mutations were transformed into Atlyk5-2 mutant plants. Eight individual transgenic plants were used for this measurement. Data are mean ± SE. Asterisks indicate significant difference relative to H2O treated Col-0 wild-type plants. (p < 0.01, Student's t test). (D) Chitin binding affinity of AtLYK5 and AtLYK5 mutant proteins as labeled in (C) detected by anti-HA antibody. Upper panel shows input of each transgenic plant, lower panel shows western blot after pull down with chitin-magnetic beads.