Abstract
Spinal muscular atrophy (SMA) is a leading genetic cause of infant mortality. The disease originates from low levels of SMN protein due to deletion and/or mutations of SMN1 coupled with the inability of SMN2 to compensate for the loss of SMN1. While SMN1 and SMN2 are nearly identical, SMN2 predominantly generates a truncated protein (SMNΔ7) due to skipping of exon 7, the last coding exon. Several avenues for SMA therapy are being explored, including means to enhance SMN2 transcription, correct SMN2 exon 7 splicing, stabilize SMN/SMNΔ7 protein, manipulate SMN-regulated pathways and SMN1 gene delivery by viral vectors. This review focuses on the aspects of target discovery, validations and outcome measures for a promising therapy of SMA.
Spinal muscular atrophy: genetics & phenotype
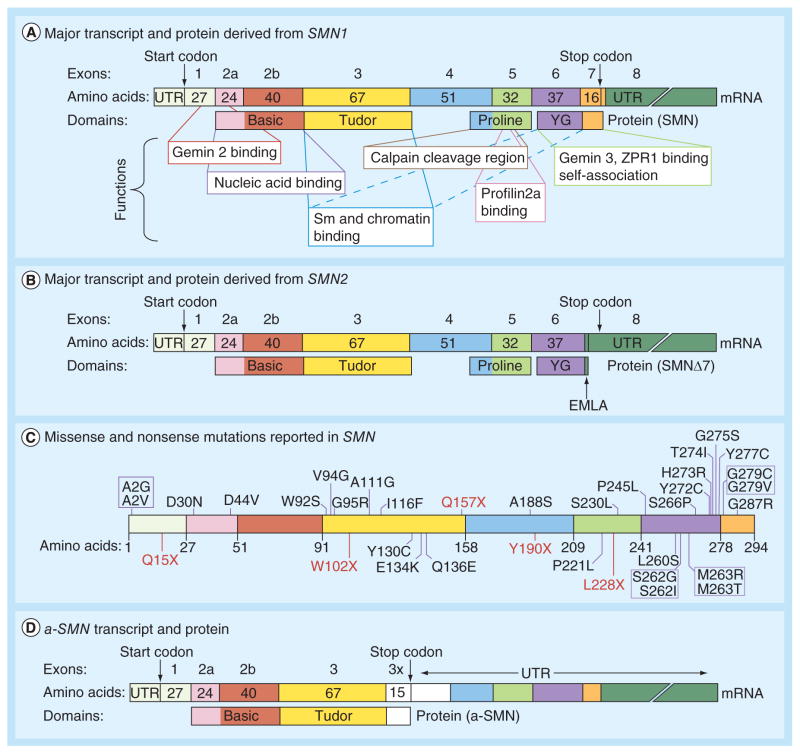
Spinal muscular atrophy (SMA) is a neurode-generative disease and a leading cause of infant mortality that occurs in approximately 1 in every 10,000 live births [1]. The disease manifests as progressive degeneration of α motor neurons in the spinal cord and the consequent atrophy of muscles. In >95% of cases, SMA is caused by the deletion, mutation or gene conversion of SMN1, a gene that encodes the ubiquitously expressed SMN (Figure 1A) [2,3]. SMN2, a nearly identical copy of SMN1, fails to compensate for the loss of SMN1 due to a C to T substitution (C6U in the mRNA) at the sixth position of exon 7 and an A to G substitution (A100G) at the 100th position of intron 7. The combination of C6U and A100G mutations triggers massive skipping of SMN2 exon 7 [4,5]. The exon 7-skipped transcript encodes SMNΔ7, a truncated protein that is partially functional and quickly degraded [6,7].
Figure 1. Description of transcripts and proteins encoded SMN.
(A) Diagrammatic representation of the major transcript and the full-length protein derived from SMN1. Exons are presented as colored boxes with the number of amino acids noted within each box. Arrows indicate locations of the start and stop codons. Corresponding protein domains and their functions are indicated. The presentation is adapted from [8]. The new Tudor domain function of chromatin binding was recently reported [9]. (B) Diagrammatic representation of the major transcript and the truncated protein derived from SMN2. Arrows indicate locations of the start and stop codons as well as EMLA degron that renders the protein unstable. Other details including color codes are the same as in (A). (C) Diagrammatic presentation of SMN protein with the mutations reported to date. Amino acids are represented by their one-letter codes. Position and type of mutations are indicated, with missense mutations shown in blue and nonsense mutations shown in red. Other details including color codes are the same as in (A). (D) Diagrammatic representation of a-SMN transcript and protein. Presence of an extra exon due to inclusion of a portion of intron 3 (3×) is shown as white box. Other details including color codes are the same as in (A).
UTR: Untranslated region.
SMA patients show remarkable variability in phenotypes. The spectrum ranges from fetuses that die in utero or soon after birth (type 0), infants born noticeably afflicted who die within 2 years of birth (type I), patients able to sit upright but not walk and who survive into their teens and adulthood (type II), patients able to walk independently with a normal or near normal lifespan (type III) and patients with adult-onset progressive muscle weakness (type IV). Barring a few exceptions [1,10–11], the copy number of SMN2 appears to correlate at least partially with SMA disease severity [12–14]. Increased SMN2 copy number may result from increased conversion of SMN1 to SMN2 or duplication of SMN2 [15,16]. SMN2 does generate low levels of full-length transcript capable of producing enough SMN for the majority of cell types, except motor neurons and muscles, to survive [17]. There is an ongoing search for tissue and cell-type specific regulators that are impacted by low levels of SMN. Several recent reviews describe progress on various fronts towards SMA therapy [10,18]. Based on independent reports of antisense oligonucleotide (ASO)- and gene-therapy-based preclinical studies, it is increasingly evident that postnatal treatment aimed at upregulation of SMN can substantially increase the lifespan of mouse models of severe SMA [18]. In this review, we provide an account of regulatory mechanisms that could be exploited to accelerate the therapeutic development for SMA.
Structure–function relationship in SMN
SMN is a multifunctional protein coded by eight exons: 1, 2a, 2b, 3, 4, 5, 6 and 7; exon 8 is not translated (Figure 1A). The SMN protein contains several functional domains including Gemin2-binding, nucleic acid binding, Tudor, self-association and cal-pain cleavage (Figure 1A) [8,9]. The interaction of SMN with Gemin2 has been suggested to be important for several SMN functions including small nuclear ribonucleoprotein (snRNP) biogenesis [19], signal recognition particle biogenesis [20], DNA recombination [21], motor neuron trafficking of mRNAs [22] and translation regulation [23]. Point mutations across the entire primary structure of the protein have been linked to SMA pathogenesis (Figure 1C; Supplementary Table 1) [1–3,11,24–55]. None of the deletions or missense mutations of SMN produce toxic effects and/or generate protein aggregates. Therefore, loss of SMN function associated with deletions or missense mutations is considered to be the major cause of SMA.
Skipping of exon 7 replaces the sixteen C-terminal amino acid residues of exon 7 with four amino acids (EMLA) coded by exon 8 (Figure 1B). EMLA serves as a degradation signal in SMNΔ7 [7]. Compounds that allow read-through of the first stop codon within exon 8, including a neomycin derivative, were shown to stabilize SMNΔ7 and may potentially provide a therapeutic option for SMA (Figure 2) [56]. Although not tested in SMA patients, the read-through compound ataluren has shown successful results in patients with Duchenne muscular dystrophy [57] and is undergoing Phase III clinical trials.
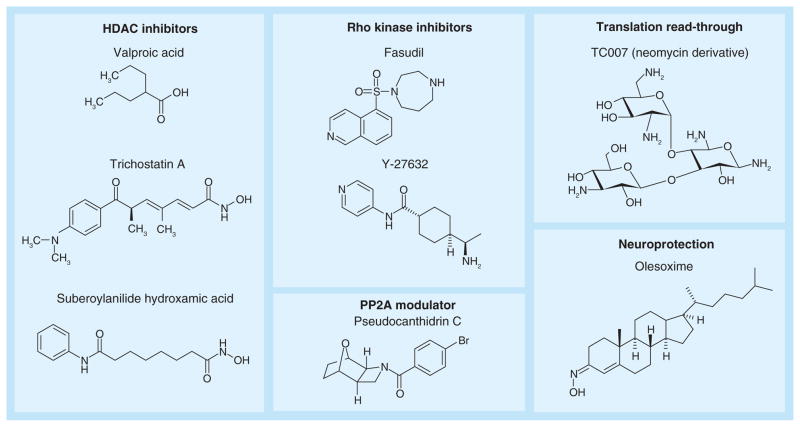
Figure 2. Select small compounds that have been investigated for spinal muscular atrophy therapy.
Select HDAC inhibitors that have been tested in mouse models of spinal muscular atrophy (SMA) and in the case of valproic acid in several clinical trials in SMA patients [58]. Rho kinase inhibitors [59,60] and the read-through compound TC007 [56] have also been tested in mouse models of SMA. Pseudocantharidin C modulates the activity of PP2A to increase SMN2 exon 7 inclusion [61]. The neuroprotective compound olesoxime is currently being tested in a Phase II clinical trial of type II and III SMA patients [62].
One of the most well characterized functions of SMN is assembly of snRNPs [63]. Participation of SMN in snRNP assembly is mediated through the SMN complex, which is comprosed of several proteins including Gemin2–8 and unrip [64]. While snRNP assembly is a cytosolic process, SMN is also localized in nuclear bodies called gems. It remains to be seen if gems have specific functions or whether they are the signatures of an ordered nuclear organization forced by the spatial and temporal arrangement of other macromolecules. Other less studied SMN functions include detection of specific chromatin modifications [9], transcription [65,66], translation [23], signal transduction [67], stress granule formation [68] and macromolecular trafficking [69,70]. Motor neurons are particularly susceptible to the loss of SMN protein, although the reasons underlying this susceptibility are not well understood. One plausible cause of neuronal susceptibility would be loss of SMN interactions with neuron-specific proteins such as HuD, a protein implicated in trafficking of mRNAs [71,72]. For instance, it was shown that interaction of SMN and HuD with mRNA cpg15 rescues motor neuron axonal deficits [71]. A role for SMN in axonal trafficking could be critical for neuromuscular junction (NMJ) formation, neurite outgrowth, myoblast fusion and myofibril integrity [72].
The molecular mass of cellular SMN determined by western blot falls between 38 and 42 kDa, a range that is substantially higher than the predicted mass of approximately 32 kDa. This discrepancy of SMN molecular mass could be attributed to post-translational modifications (PTMs). Indeed, phosphorylation of SMN is considered to be important for snRNP assembly [73,74], and phosphorylation of SMN by protein kinase A increases SMN stability [6,75]. Conversely, small molecules that inhibit GSK-3, and presumably prevent GSK-3-mediated phosphorylation of SMN, can increase SMN stability [76]. Ubiquitination of SMN causes its degradation through the ubiquitin proteasome system [6]. The significance of other PTMs, including but not limited to glycosylation, palmitoylation, sumoylation and transamidation, has yet to be explored. A less abundant, shortened isoform of SMN, a-SMN, is generated by alternative splicing in which a portion of SMN intron 3 is included in a tissue-specific and developmentally regulated manner (Figure 1D) [77]. The a-SMN isoform is evolutionarily conserved between humans and rodents [78]. Although the physiological role of a-SMN is poorly understood, a-SMN was recently shown to promote axon growth, stimulate cell motility and regulate expression of chemokines (CCL2, CCL7) and IGF-1 [79].
SMN depletion in mice during embryonic development causes a severe phenotype with early postnatal lethality. SMN restoration can ameliorate this phenotype, but only if done soon after birth [80,81]. In Smn knockout zebrafish, SMN must be restored relatively soon after motor neuron birth for normal motor axon development [82]. SMN is particularly important in the maturation and repair of NMJs, as early depletion prevents the formation of mature NMJs and leads to early postnatal death [83]. However, mice reach an SMN-refractory period after about 2 weeks of age where SMN depletion does not lead to early postnatal lethality, but much later in life abnormal NMJs are apparent [83]. While SMN is known to affect motor neurons, muscles and possibly other tissues are also affected independent of motor neuron defects [17,84]. Overall, these studies suggest that treatments for SMA patients should be given as early as possible to increase the SMN level and potentially ameliorate any abnormalities as much as possible.
SMN pre-mRNA splicing regulation
The mechanism of SMN2 exon 7 splicing has been extensively studied since it is directly relevant to potential SMA therapies. Due to the close proximity of C6U to the 3′ splice site (3′ ss) of exon 7, early studies hypothesized that C6U weakened it. Considering that very small sequence motifs may define the specificity of RNA-protein interactions, it is not uncommon that loss of one motif due to a point mutation could be accompanied by a gain of another motif. Indeed, the inhibitory effects of C6U have been suggested to abrogate an enhancer element associated with SF2/ ASF, as well as create a silencer element associated with hnRNP A1 [85,86] (Figure 3A). Furthermore, C6U strengthened a terminal stem loop (TSL1), an inhibitory RNA structure (Figure 3B) [87]. Subsequent studies revealed a negative role of Sam68 and a positive role of hnRNP Q exerted through the 6th position of exon 7 [88]. It is now also known that splicing regulators, such as Tra2-β1, SRp30c, hnRNP G and TDP-43, stimulate exon 7 inclusion by interacting directly or indirectly with exon 7 [88]. Consequently, factors that modulate the phosphorylation of Tra2-β1 and other splicing factors, including the pseudocantharidin compounds, have been shown to affect SMN2 exon 7 splicing as well (Figure 2) [61].
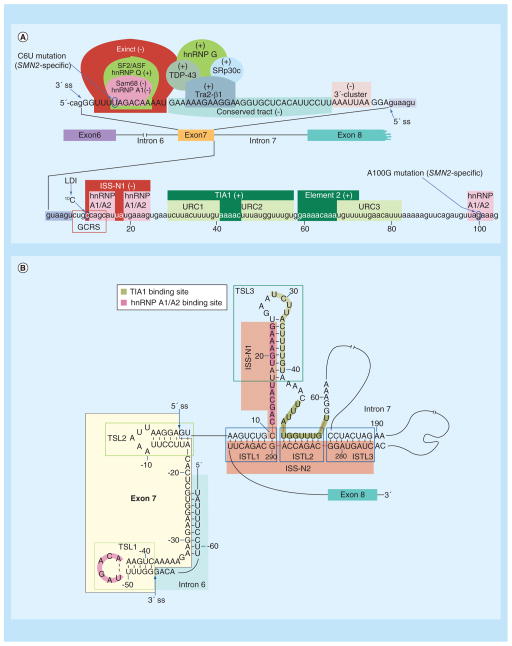
Figure 3. Regulators of SMN exon 7 splicing (see facing page).
(A) Diagrammatic representation of cis-elements and transacting factors that affect splicing of exon 7. Exon 7 sequence is shown in capital letters, while adjacent intronic sequences, including the first 104 nucleotides of intron 7, are shown in lower-case letters. The ss of exon 7 are indicated by arrows; in addition, the 5′ ss is highlighted with a blue arrow. Nucleotide numbering starts from the first position of intron 7. Cis-elements/transacting factors that promote exon 7 inclusion are indicated by (+), while cis-elements/transacting factors that promote exon 7 skipping are indicated by (−). Exinct, the conserved tract and 3′-Cluster were identified by in vivo selection of the entire exon 7 [89]. Element 2 and binding sites for hnRNP A1/A2, Sam68, SF2/ASF, hnRNP Q, Tra2-β1, TDP-43, hnRNP G and SRp30c were described previously [88,90]. TIA1 binds to URC1 and URC2 located in intron 7 and promotes exon 7 inclusion [90]. ISS-N1, along with the overlapping GCRS and the 10C, which is involved in LDI, contribute to skipping of exon 7 [90]. TSL2 structure sequesters the 5′ ss of exon 7 and promotes its skipping [91]. The presentation is adapted from [92]. (B) Schematic representation of the partial RNA secondary structure formed by the sequences that constitute SMN2 exon 7, the last 14 nucleotides of intron 6 and the entire intron 7. The structure is based on the results of chemical structure probing performed in [91,92]. Arrows indicate the 5′ and 3′ ss of exon 7. Exon 7 and intron 6 sequences are highlighted in yellow and light blue, respectively. Nucleotide numbering starts from the first position of intron 7. Three TSLs are marked by green boxes. The juxtaposed internal stems formed by LDI (ISTLs) are marked by blue boxes. ISS-N1 and ISS-N2 are highlighted in red. The adjacent 3′-strands of ISTL1, ISTL2 and ISTL3 constitute ISS-N2. Binding sites of TIA1 and hnRNP A1/A2 are indicated.
GCRS: GC-rich sequence; ISS-N1: Intronic splicing silencer N1; LDI: Long-distance interaction; ss: Splice site; TSL: Terminal stem-loop; URC: U-rich cluster.
Available algorithms do not predict the relative significance of exonic positions in splicing regulation due to variable contexts of exons. To circumvent this problem, we developed a novel in vivo selection protocol and employed it in the context of the entire exon 7 of SMN1 [89]. This complex experiment uncovered three regulatory regions: extended inhibitory context (Exinct), conserved tract and 3′-cluster (Figure 3A) [89]. Exinct and 3′-cluster are negative regions located close to the 3′ and 5′ ss of exon 7, respectively. Conserved tract is a positive region that covers most of the middle portion of exon 7 and fully encompasses the previously described GA-rich sequence associated with Tra2-β1 interaction (Figure 3A). The nature of the regulatory regions identified by in vivo selection corroborated well with the results of an independent study in which overlapping ASOs were used to probe the significance of regulatory cis-elements within the entire exon 7 in the context of endogenous SMN2 [93]. The identification of Exinct, and its further validation by an ASO-based approach, lends additional support to the hypothesis that C6U strengthens a negative cis-element within SMN2 exon 7.
In recent years, SMA patients with unique mis-sense mutations within exon 7 of SMN1 and SMN2 have been reported (Supplementary Table 1). One such example is a mild form of SMA associated with a G to T mutation at the 29th position (G29U in the mRNA) of SMN1 exon 7 [53]. Although not yet proven, it is expected that G29U would likely trigger SMN1 exon 7 skipping due to its positioning within the conserved tract that spans from the 16th to 44th positions of exon 7. It is also possible that G29U creates an inhibitory cis-element. Another less severe form of SMA is associated with the loss of SMN1 coupled with a G to C mutation at the 25th position (G25C) of SMN2 exon 7 [48,51]. This is one of the rare examples in which deletion of SMN1 is partially rescued by a single nucleotide mutation within SMN2 exon 7. G25C moderately stimulated SMN2 exon 7 inclusion in the context of a minigene. Based on in vitro binding assays, it was argued that G25C abrogates a composite regulatory element associated with hnRNP A1 and SF2/ ASF [51]. It is still unknown if G25C creates a novel enhancer and/or brings structural changes favorable for SMN2 exon 7 inclusion. Of note, G25C replaces a hydrophobic residue (glycine) with a charged hydrophilic residue (arginine) with the potential to alter the properties of SMN. However, the contribution of this mutant SMN to modulation of SMA severity requires further examination.
In addition to the defining C6U and A100G mutations of SMN2, the weak 5′ ss of exon 7 serves as the limiting factor for SMN2 exon 7 inclusion. This was first revealed by the results of our in vivo selection of the entire exon 7; a non-wild type G residue was overwhelmingly selected at the last position of exon 7 [89]. The presence of a G residue at this exonic position extends the base pairing between the 5′ ss of exon 7 and U1 snRNA, an essential component of U1 snRNP that defines the 5′ ss of an exon. This finding clearly supported the hypothesis that a poor recruitment of U1 snRNP at the 5′ ss is the cause of SMN2 exon 7 skipping. This hypothesis was validated when a mutant U1 snRNA that increased the base pairing with the 5′ ss of exon 7 robustly stimulated SMN2 exon 7 inclusion [91]. Subsequent reports revealed a number of inhibitory cis-elements in the vicinity of the 5′ ss of exon 7. These inhibitory elements include TSL2, a 15-nucleotide long intronic splicing silencer N1 (ISS-N1) and an 8-nucleotide long GC-rich sequence (GCRS) that partially overlaps with ISS-N1 (Figure 3A) [91,94–95]. It has been shown that the last 14 residues of ISS-N1 encompass two putative binding sites of hnRNP A1/A2 (Figure 3) [96]. The first residue of ISS-N1 is a cytosine that occupies the 10th position (10C) of SMN intron 7 and participates in an inhibitory long-distance interaction (LDI) involving downstream intronic sequences [92,97]. Given the negative role of ISS-N1 and GCRS, it is not surprising that ASO sequestration of these motifs promoted SMN2 exon 7 inclusion [8,94–95].
TIA1 and TIAR stimulated SMN2 exon 7 inclusion by binding to intronic U-rich clusters (URC1 and URC2) downstream of ISS-N1 (Figure 3A) [90]. Recently, a missense mutation within TIA1 was linked to Welander distal myopathy, a late onset autosomal dominant disease [98]. Muscle biopsies from Welander distal myopathy patients carrying the TIA1 mis-sense mutation showed elevated levels of SMN2 exon 7-skipped transcripts [98]. This is the first evidence that demonstrated the critical role of a SMN exon 7-splicing-associated factor in the context of a human disease. This finding clearly underscores the likely consequences of aberrant expression of a splicing factor on the severity of SMA. Since TIA1 is a Q-rich domain containing protein, we suspect that other RNA-binding proteins with similar domain organization would affect SMN2 exon 7 splicing and SMN levels and consequently SMA severity. It should be mentioned that TIA1 is a critical component of stress granules, formation of which is an important cellular process that is negatively impacted by low levels of SMN [68]. Therefore, the impact of TIA1 on the severity of SMA is likely to be compounded by several TIA1-associated functions.
Recently, we interrogated folding of SMN2 intron 7 and identified several regulatory structures, including TSLs and internal stems formed by LDIs (ISTLs) (Figure 3B & Figure 4) [92]. In particular, we identified ISTL1, a unique structure in which the 5′ and the 3′ strands of a RNA–RNA duplex are separated by more than 250 nucleotides. Interestingly, 10C and half of GCRS are locked in ISTL1 (Figure 3B). Although the significance of ISTL1 is not yet fully understood, it represents an example of a structure generated by a unique folding of the entire SMN2 intron 7 (Figure 3). The continuous sequence stretch encompassing the 3′ strands of ISTL1, ISTL2 and ISTL3 constitute ISS-N2, an inhibitory region in the middle of intron 7 (Figure 3).
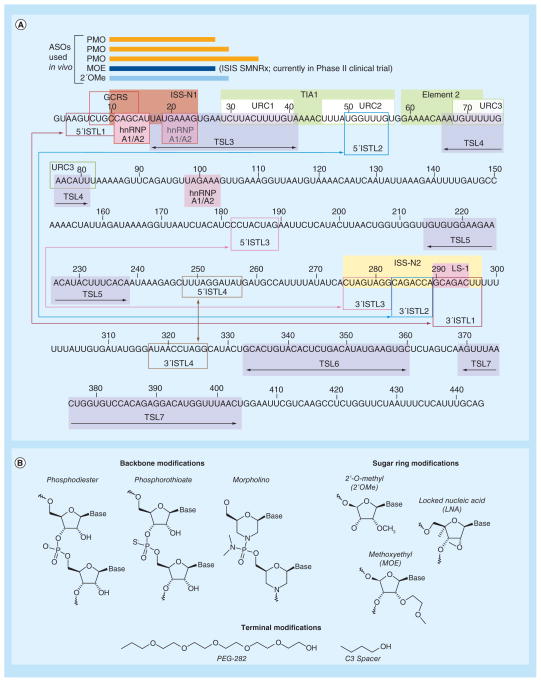
Figure 4. Linear representation of the entire SMN2 intron 7 (see facing page).
(A) Colored boxes designate the structural elements and splicing cis-elements. Numbering starts from the first position of intron 7. Double arrow within a given box indicates a TSL, whereas, a double arrow outside of boxes in the corresponding color shows the 5′ and 3′ strands of a particular ISTL. ISS-N1-targeting ASOs used for in vivo studies are shown as bars. The linear structure is adapted from [92]. (B) Chemical structures for a ASO backbone (unmodified phosphodiester, phosphorothioate and morpholino) and sugar ring modifications (2′-O-methyl, 2′-methoxyethyl and locked nucleic acid). The terminal modifications shown, PEG-282 and C3 spacer, were successfully utilized to increase the in vivo stability of a short ASO [99].
ASO: Antisense oligonucleotide; GCRS: GC-rich sequence; ISTL: Internal stem formed by LDI; LDI: Long-distance interaction; TSL: Terminal stem-loop; URC: U-rich cluster.
Antisense-based therapy
In recent years, a wide range of research has utilized ASO-based approaches to modulate transcription, splicing and translation [100,101]. A variety of ASO-based strategies have also been adopted to stimulate SMN2 exon 7 inclusion, albeit with varying degrees of success as recently reviewed in [102,103]. Discovery of ISS-N1 in 2006 provided a major breakthrough for ASO-mediated splicing correction in SMA [94]. ISS-N1 earned the name of a ‘master checkpoint’, since deletion of ISS-N1 or an ASO-mediated sequestration of ISS-N1 fully restored SMN2 exon 7 inclusion even in the absence of a number of positive regulatory elements [104]. Advantageously, an ASO that anneals to an intronic sequence does not interfere with the translation and transport machinery. Therefore, ISS-N1 serves as an ideal target for the restoration of the full-length protein. Indeed, very low concentrations of an ISS-N1-targeting ASO significantly increased levels of SMN in SMA patient cells [94].
Since the discovery of ISS-N1, several independent studies with ISS-N1-targeting ASOs have been performed, the details of which have been recently reviewed [102]. These studies have employed ASOs with different modifications, including 2′-O-methyl coupled with uniform phosphorothioate backbone, 2′-O-methoxy-ethyl (MOE) coupled with uniform phosphorothioate backbone and phosphorodiamidate morpholino oligomers (Figure 4B) [102]. Of particular note is a study from the Krainer group that showed approximately 25-fold increase in lifespan of severe type I Taiwanese mice upon early subcutaneous administration of an ISS-N1 18-mer MOE ASO [105]. While this strategy resulted in the longest survival increase of any treatment performed in this model, the effect was less promising when the MOE ASO was delivered through intracerebroventricular (i.c.v.) route, although the i.c.v. dose was substantially lower than subcutaneous administration [105]. On the contrary, independent studies conducted with ISS-N1-targeting phosphorodiamidate morpholino oligomers have shown very promising results for i.c.v. administration in both type I Taiwanese and Δ7 SMA mouse models of SMA [106–108]. These results also underscore the need for the optimization of ASO size, since a difference of a few nucleotides may dramatically impact the in vivo efficacy of an ASO. Furthermore, given that SMA patients exhibit deficits in tissues outside the nervous system [17,84], it appears that treatment should reach the nervous system and peripheral tissues. Currently, the MOE ASO targeted against ISS-N1 (ISIS-SMNRx) is recruiting patients for a Phase II clinical trial [109] after the successful completion of Phase I clinical trial [110].
GCRS and ISS-N2 represent additional antisense targets for ASO-mediated splicing correction in SMA (Figure 4A). Located within SMN2 intron 7, these unique targets offer different sequence compositions and mechanisms for promoting SMN2 exon 7 inclusion. The GCRS target in particular provides a rare advantage of utilizing a short ASO to increase SMN2 exon 7 inclusion. There is a general misconception that a small ASO is bound to be less specific due to preponderance of similar motifs throughout the genome/ transcriptome. This misconception is built on an unproven hypothesis that a large ASO does not tolerate mismatched base pairs. Based on the bifunctional ASOs and seed interactions of miRNAs that do not require full complementarity with the target [111,112], large ASOs can function while tolerating more than 50% mismatched base pairs. On the other hand, small ASOs show limited or no tolerance for a single mismatch base pair. Indeed, tiny ASOs targeting seed sequences of microRNAs have shown commendable efficacy in vivo [113]. Furthermore, a recent report validated the therapeutic efficacy of a GCRS-targeting short ASO in two severe mouse models of SMA [99]. Therefore, employment of a small ASO as a potential therapy of SMA remains an attractive proposition.
Transcriptional & epigenetic regulation
Upregulation of SMN2 transcription could potentially increase SMN without altering the splicing pattern of SMN2 exon 7. The human SMN1 and SMN2 gene promoters are nearly identical in sequence and activity [114,115]. While the principal transcription initiation site (TIS) is located 163 base pairs upstream of the translation initiation site, a second TIS has been mapped 246 base pairs upstream and appears to be used during fetal development [116]. Sequences up to 4.6 kilobases upstream of the TIS have been implicated in transcriptional regulation of SMN1/ SMN2 and harbor putative bindings sites of transcription activators including CREB and ELK-1 [117,118].
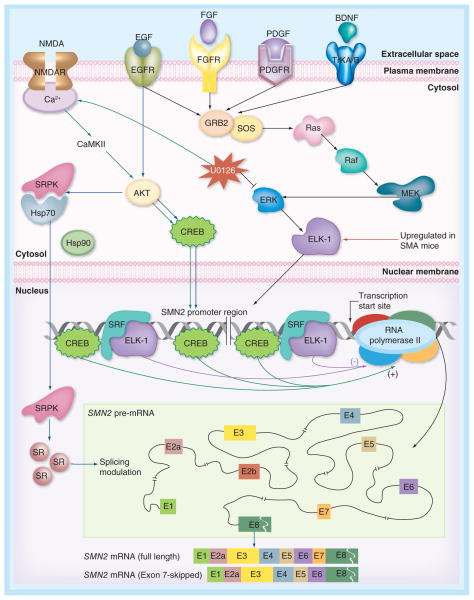
There are very limited studies on the molecular mechanism by which transcription of SMN2 exon 7 is regulated. Prolactin, a polypeptide hormone and an activator of the JAK2/STAT5 signaling pathway, increased SMN2 transcription and SMN in both human neuron-committed teratocarcinoma (NT2) cells and the Δ7 SMA mouse model and improved the disease phenotype of Δ7 SMA mice [119]. Inhibition of the MEK/ERK pathway coupled with the activation of the AKT/CREB pathway is another mechanism recently shown to enhance SMN2 transcription (Figure 5) [118,120]. Oral treatment with selumetinib, a MEK/ERK inhibitor currently in Phase II clinical trials [121], significantly enhanced expression of SMN2 in spinal cord and increased lifespan by approximately 3.4-fold in a severe SMA mouse model [118]. Similar results were obtained with intrathecal administration of U0126, an inhibitor of the MEK/ERK/Elk-1 signaling pathway [118]. While the MEK/ERK/ELK-1 and the PI3K/AKT/CREB signaling pathways are known to cooperate with beneficial consequence to neurons, it appears that these two pathways have opposing roles in transcriptional regulation of SMN2 in the spinal cord of type I SMA-like mice. The ERK/ELK-1 pathway becomes constitutively overactive in the spinal cord of SMA-like mice [120], exerting a negative effect on expression (transcription) of SMN2 [118]. CREB is a positive regulator of SMN transcription [122], whereas ELK-1 plays a repressive role on transcription in certain circumstances [123]. In the spinal cord of type I SMA-like mice, inhibition of ELK-1 phosphorylation through MEK/ERK pathway inhibition by U0126 leads to activation of the AKT/CREB pathway. The crosstalk between MEK/ERK and AKT/CREB pathways is facilitated by CaMKII (Figure 5) [118]. Activation of spinal cord NMDA receptor that induces calcium flux and consequently activates AKT/CREB pathway, enhances SMN2 gene expression in severe SMA mice [120]. Interestingly, massive reprogramming of alternative splicing in response to EGF signaling has been reported [124]. For instance, it has been shown that activation of AKT induces autophosphorylation of SRPKs, an action that enhances SRPK translocation and SR protein phosphorylation [124]. Therefore, it is likely that the increase in SMN2 transcript levels in response to activation of JAK2/STAT5 and/or AKT/CREB pathway is partly due to an increase in the rate of splicing.
Figure 5. Signaling pathways implicated in regulation of SMN2 transcription and SMN2 exon 7 splicing.
Simplified version of intracellular MEK/ERK and AKT/CREB signaling pathways that affect SMN transcription, including the upstream regulators (NMDA, EGF, FGF, PDGF and BDNF). Activation of spinal cord NMDA receptors or inhibition of ERK by U0126 activates the AKT pathway leading to increased SMN2 transcription [118,120]. Activation of AKT triggers autophosphorylation of SRPK followed by its nuclear import and consequent phosphorylation of SR proteins that regulate alternative splicing [124]. Binding sites of transcription factors within SMN2 promoter are shown. In the inset for SMN2 pre-mRNA, exonic (coding) and intronic (non-coding) regions are indicated by boxes and lines, respectively. Two major SMN2 splice isoforms are shown at the bottom.
EGFR: EGF receptor; FGFR: FGF receptor; NMDAR: NMDA receptor; PDGFR: PDGF receptor.
Histone acetylation and deacetylation are important epigenetic mechanisms by which transcription is activated and repressed, respectively. A number of HDAC inhibitors have been found to increase transcription of SMN2, including suberoylanilidehydroxamic acid, trichostatin A, and US FDA approved drugs hydroxy-urea and valproic acid (Figure 3) [58]. However, clinical trials conducted with valproic acid and hydroxyurea did not show promising results [125–129]. It is possible that HDAC inhibitors would exhibit greatest efficacy if administered immediately to newborns or shortly after birth, although patients this young would be difficult to enroll in clinical trials. Perhaps HDAC inhibitors could be utilized as adjunct therapy, especially for patients with more complicated manifestations of SMA.
RNA decapping
Decapping is a regulated mRNA scavenging process catalyzed by the enzyme DcpS, a member of the HIT family of pyrophosphatases [130]. Quinazolines are a family of DcpS inhibitors that were identified as SMN modulating compounds in a high-throughput screening [131]. In order to improve the delivery across the blood–brain barrier, several quinazoline derivatives were developed, including a promising candidate known as RG3039. Daily intraperitoneal administration of RG3039 improved NMJ function and modestly extended lifespan of Δ7 SMA mice [132]. Similar results were obtained in Taiwanese type I mice that received daily oral administration of RG3039; RG3039 also significantly extended median lifespan from 18 to 112 days in the intermediate Smn2B/− mouse model [133]. Currently, RG3039 is undergoing Phase I clinical trial.
Gene therapy of SMA
Gene therapy as a treatment for SMA could potentially cure the disease through the delivery of SMN1 and consequent restoration of SMN. The first gene therapy study reported in 2004 was conducted with a lentiviral vector to deliver SMN1 gene in the Δ7 SMA mice [134]. The results were modest; treated Δ7 mice survived merely 3 days longer than the usual 14 days, but nevertheless it was a promising result at the time. Over the past several years, adeno-associated vectors (AAV) class 8 and 9 have emerged as potential vehicles for delivering genes to the CNS [135,136] and have been tested in mouse models of SMA. i.c.v. administration at birth of SMN1 cDNA packaged in AAV8 and scAAV8 vectors markedly increased the median lifespan of the mice to 57 and 157 days, respectively, and both vectors improved neuromuscular architecture and motor function [137]. The majority of gene therapy studies in SMA mouse models, however, have involved scAAV9. Intravenous (iv.) administration of scAAV9 carrying SMN1 cDNA at postnatal day 1 restored endplate current at neuromuscular junctions and extended lifespan of Δ7 SMA to at least 250 days [138]. The same gene therapy protocol rescued the severe bradycardia and improved electrocardiographic features [139] and the heart structural defects [140] observed in untreated Δ7 SMA mice. Recent studies utilized codon optimized SMN1 cDNA in scAAV9 with different promoters. Iv. administration of SMNopti under control of the phosphoglycerate kinase promoter spared motor neurons from death, prevented motor function deficits and increased median survival to 199 days (compared with 13 days for untreated mice) [141]. However, iv. administration of codon-optimized SMN1 under cytomegalovirus control only extended lifespan up to 70 days [142]. Findings from these reports suggest the importance of the promoter in addition to the SMN1 sequence. However, the efficacy of route of viral administration remains unclear. Although equal efficacies have been reported for iv., i.c.v. and intramuscular deliveries of scAAV9-SMN1 [143,144], the requirement of high viral titer (up to 1014 viral genomes) may limit the clinical utility of peripheral administration. Additional studies in different mouse models of SMA, as well as toxicology studies in non-human primates, are needed for gene therapy to be an attractive option in SMA. Recently, the FDA approved a Phase I clinical trial for systemic AAV9 delivery of SMN1 [145].
Disease modifiers of SMA
Several genes, including NAIP, H4N4 and PLS3 are known to affect SMA severity [146,147]. NAIP and H4N4 genes are located in close proximity to SMN1 on human chromosome 5. The most severe type I patients are found to have deletions of NAIP and/or H4N4 in addition to deletion of SMN1. The PLS3 gene is located away from the SMN locus on human chromosome 10 and encodes the F-actin bundling protein plastin-3 (PLS3). Thus far, PLS3 is the only reported natural disease modifier of SMA because individuals with high PLS3 levels have been found to be protected from the harmful effects of homozygous SMN1 deletions that cause SMA [146]. Several disease modifiers have been implicated based on studies in cell-based and animal models. For instance, altered phosphorylation of Rho kinase-dependent targets that influence polymerization of actin have been suggested to play an important role in SMA pathogenesis [52]. Consistently, treatment of an intermediate mouse model of SMA (Smn2B/−) with Y-27632 or fasudil, both inhibitors of Rho kinase (Figure 2), increased survival and myofiber size [59,60]. Based on a recent transcriptome-wide analysis of motor neurons and surrounding glial cells, multiple genes, including agrin (required for NMJ maintenance), C1q (synapse pruning-promoting complement factor) and Etv1/ER81 (transcription factor for establishing sensory motor circuitry), are likely to be modifiers of SMA [148]. Dysregulation of these genes further underscores the importance of SMN in maturation of NMJs. In addition, zinc-finger protein, ZPR1, which is required for nuclear accumulation of SMN, is down-regulated in SMA patients [149]. In vitro, overexpression of ZPR1 in SMN-deficient spinal cord neurons from SMA mice stimulated neurite growth and rescued axonal growth deficits [149]. Targeting these modifying factors may provide avenues for the development of adjunct therapy for SMA. Cellular levels of these factors may also serve as markers of outcome measures for the development of SMA therapy.
Future perspective
The past 15 years have witnessed a remarkable progress in our understanding of SMA pathogenesis, SMN function, SMN transcription and SMN exon 7 splicing. Several in vivo studies have been performed for potential therapeutic compounds identified by available chemical libraries. Similar progress has taken place for the therapeutic development of other genetic diseases. However, genuine progress towards therapy in human patients has been less than impressive due to the lack of reproducibility of most of the in vivo studies performed using mouse models. According to a recent report, the minimum cost of testing the efficacy of a single dose of a given compound in a mouse model of amyotrophic later sclerosis (ALS), a disease similar to SMA, comes out to be approximately US$330,000 [150]. This report suggests a minimum time requirement of 9 months to realistically conclude a meaningful preclinical study in ALS. Significantly, the report also provides a clue why most claims based on low-budget preclinical studies in ALS were not reproducible and eventually failed in clinical trials [150]. The findings of this report are instructive for both investigators and funding agencies (public, private and non-profit organizations) in factoring the realistic cost and time of pursuing a preclinical study in an animal model. Ideally, an in vivo study is warranted only after rigorous cell-based experiments have validated the efficacy and target specificity of a compound.
SMA is a disease with the potential to be cured by correction of SMN2 exon 7 splicing. Compounds that correct SMN2 exon 7 splicing should be rigorously evaluated for the specificity, particularly at higher concentrations. Similarly, the mechanism of compounds that increase SMN levels independent of SMN2 exon 7 splicing should be stringently evaluated for specificity. Better solubility (in aqueous solutions) and high activity at low concentrations in cell-based studies is a good indicator if a compound is suitable for in vivo application. The manner in which ISS-N1 target was discovered and subsequently validated in the cell-based assays led to an overwhelming interest in an ASO-based therapy for SMA. Consequently, ISS-N1 emerged as the natural contender for testing the efficacy of different ASO chemistries in various mouse models of SMA. While several independent reports on ISS-N1-targeting ASOs have shown unprecedented therapeutic benefits in vivo, rigorous and independent evaluations of different chemistries with a substantially large number of animals per treatment group over a long period of time are yet to be conducted. Gender variations should also be factored in the experimental design. Gene therapy is another promising approach for the treatment of SMA, particularly if DNA coding for SMN is safely delivered to all needed tissues at the very early stages of development. Herein, preclinical studies should be conducted with a larger cohort of animals considering variability in administration and tolerability of individual animals.
Progress in the field of ASO-based and gene therapies does not in any way diminish the need for other approaches, particularly small compounds that could serve as major means for therapy at the advanced stages of the disease or as an adjunct therapy to gene-based treatments. The most important lesson to be learnt from the journey of the therapeutic development for SMA is to continue to uncover the mechanism of the disease at the molecular, cellular and organismal level. There are many more therapeutic targets hidden in the sequences and structures of molecules that affect SMN2 transcription, SMN2 exon 7 splicing, SMN stability and SMN-dependent processes.
Supplementary Material
Executive summary.
Spinal muscular atrophy: genetics & phenotype
Spinal muscular atrophy (SMA) is the leading genetic cause of infant mortality that occurs in approximately 1 in every 10,000 live births. SMA patients show a broad spectrum of phenotype with type 0 being the most severe and type IV being the mildest.
Humans carry two near identical copies of SMN gene: SMN1 and SMN2. SMA is caused by the loss of SMN1 coupled with the inability of SMN2 to compensate for the loss of SMN1.
The major problem with SMN2 gene is exon 7 skipping that produces SMNΔ7, a truncated SMN that is rapidly degraded.
Structure–function relationship in SMN
SMN is a multifunctional protein coded by eight exons. The functional domains present within SMN include Gemin2-binding, nucleic acid binding, Tudor, self-association and calpain cleavage. Point mutations across the entire primary structure of the protein have been linked to SMA pathogenesis.
Interaction of SMN with Gemin2 is important for snRNP biogenesis, signal recognition particle biogenesis, translation regulation, DNA recombination and motor neuron trafficking of mRNAs.
Regulation of SMN2exon 7 splicing
SMN2-specific C6U and A100G substitutions cause massive skipping of SMN2 exon 7. C6U and A100G substitutions create binding motifs for the inhibitory factor hnRNP A1/A2. Depletion of hnRNP A1/A2 promotes SMN2 exon 7 inclusion.
In vivo selection of the entire SMN exon 7 revealed a weak 5′ ss as the limiting factor for SMN exon 7 inclusion. The inhibitory cis-elements weakening the 5′ ss of SMN exon 7 include terminal stem loop 2, intronic splicing silencer (ISS)-N1, GC-rich sequence (GCRS) and ISS-N2.
ISS-N2 participates in the formation RNA structures (ISTL1, ISTL2 and ISTL3) facilitated by long-distance interactions. ISTL1 is a rare structure in which two stems of a RNA–RNA duplex is separated by more than 250 nucleotides.
TIA1 regulates SMN2 exon 7 splicing by binding to intronic sequences immediately downstream of ISS-N1. Pathogenic TIA1 mutation in Welander distal myopathy has been found to promote SMN2 exon 7 skipping
Antisense-based therapy
Antisense oligonucleotides targeting ISS-N1, GCRS and ISS-N2 fully restore SMN2 exon 7 inclusion and substantially elevate levels of full-length SMN in SMA patient cells. GCRS provides a unique advantage of utilizing a very small ASO for SMA therapy.
Among various oligonucleotide chemistries tested, ISS-N1-targeting phosphorodiamidate morpholino oligomers produced the best life extension benefits when delivered through the clinically relevant intracerebroventricular route in two independent studies carried with different severe mouse models of SMA.
The 2′-O-methoxyethyl ASO (ISIS-SMNRx) targeted against ISS-N1 has successfully completed the Phase I clinical trial by ISIS Pharmaceuticals (CA, USA). Patients are being recruited for a Phase II clinical trial of ISIS-SMNRx.
Transcriptional & epigenetic regulation
Upregulation of SMN2 transcription produces increased levels of SMN despite no apparent change in the splicing pattern of SMN2 exon 7. Prolactin, an activator of the JAK2/STAT5 pathway, improves the disease phenotype of a severe SMA mouse model by increasing SMN2 transcription.
Inhibition of the MEK/ERK pathway coupled with the activation of the AKT/CREB pathway enhances SMN2 transcription. Oral treatment with selumetinib, a MEK/ERK inhibitor currently in Phase II clinical trials, significantly increased lifespan of a severe SMA mouse model. A number of HDAC inhibitors have been found to increase transcription of SMN2. However, an effective HDAC inhibitor for SMA therapy is yet to be found.
RNA decapping
Quinazolines are RNA-decapping inhibitors shown to upregulate SMN. RG3039, an improved version of quinazoline, is currently undergoing Phase I clinical trial.
Gene therapy
Gene therapy offers a permanent solution to SMA if DNA coding for SMN is safely delivered to the right tissues at the right time. AAV9-based delivery of DNA coding for SMN has shown the best promise in various preclinical studies in mouse models of SMA.
Disease modifiers
Genes including NAIP, H4N4 and PLS3 have been known to modify the severity of SMA. Additional SMA modifiers have been suggested based on studies in mouse models of SMA. Targeting these modifying factors may provide avenues for the development of adjunct therapies of SMA.
Future perspective
ASO- and gene-therapy-based approaches have shown the best outcome measures in terms of extension of life span of mouse models of SMA.
Small compounds continue to be attractive agents of SMA therapy due to their ease of administration.
To uncover more promising therapeutic targets, research should continue to focus on understanding the mechanism of the disease at the molecular, cellular and organismal level.
Acknowledgments
The authors wish to acknowledge J Seo and EW Ottesen for the valuable comments on the manuscript.
Key Terms
- Antisense oligonucleotide
Antisense oligonucleotides (ASOs) are nucleic acid molecules that anneal to specific RNA or DNA sequences. ASOs have been used to block regulatory sequences to modulate alternative pre-mRNA splicing
- Small nuclear ribonucleoprotein
Small nuclear ribonucleoproteins are the building blocks of spliceosome, which catalyzes the removal of intronic sequences during the essential process of pre-mRNA splicing. SMN is involved in biogenesis of small nuclear ribonucleoproteins
- Intronic splicing silencer N1
15-nucleotide long intronic splicing silencer located from the 10th to 24th positions of SMN intron 7. Intronic splicing silencer N1 is the leading target for an ASO-mediated splicing correction in spinal muscular atrophy
- GC-rich sequence
8-nucleotide long intronic splicing silencer located from the 7th to 14th positions of SMN intron 7. GC-rich sequence is the smallest validated target for an ASO-mediated splicing correction in spinal muscular atrophy
- Intronic splicing silencer N2
23-nucleotide long deep intronic sequence located from the 275th to 297th positions of SMN intron 7. ISS-N2 is a recently validated target for an ASO-mediated splicing correction in spinal muscular atrophy
- ISIS-SMNRx
ISIS-SMNRx is the first antisense drug to undergo clinical trial for the treatment of spinal muscular atrophy. Drug is based on proprietary 2′-O-methoxyethyl chemistry of ISIS Pharmaceuticals and targets ISS-N1 sequence within SMN2 intron 7
- Quinazolines
Quinazolines are small compounds that stabilize mRNAs by inhibiting the action of decapping enzyme DcpS. RG3039 is a quinazoline derivative that has been shown to extend the lifespan of spinal muscular atrophy mice
- Adeno-associated vectors
Adeno-associated vectors have emerged as the powerful tool to deliver coding sequence for making full-length SMN in the CNS. Recently, US FDA has approved a Phase I clinical trial for AAV9 delivery of SMN1 cDNA into spinal muscular atrophy patients
- Transcriptome-wide analysis
Transcriptome-wide analysis requires high throughput sequencing of the entire transcripts from a specific cell or tissue type. Recent transcriptome-wide analyses of neuronal tissues from spinal muscular atrophy mice have revealed novel hallmarks of spinal muscular atrophy pathogenesis
Footnotes
For reprint orders, please contact reprints@future-science.com
To view the supplementary data that accompany this paper please visit the journal website at: www.future-science.com/10.4155/FMC.14.63
Financial & competing interests disclosure
This work was supported by grants from National Institutes of Health (NS055925, NS072259 and NS080294) and Sals-bury Endowment (Iowa State University, IA, USA) (to RNS). Intronic splicing silencer-N1 target (US7838657) was discovered in the Singh lab at UMASS Medical School (MA, USA). Inventors, including RNS, NNS and UMASS Medical School, are currently benefiting from licensing of intronic splicing silencer-N1 target to ISIS Pharmaceuticals. Iowa State University holds intellectual property rights on GC-rich sequence and intronic splicing silencer-N2 targets. Therefore, inventors including RNS, NNS and Iowa State University could potentially benefit from any future commercial exploitation of GC-rich sequence and intronic splicing silencer-N2. The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Prior TW. Spinal muscular atrophy diagnostics. J Child Neurol. 2007;22(8):952–956. doi: 10.1177/0883073807305668. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre S, Bürglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 3.McAndrew PE, Parsons DW, Simard LR, et al. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60(6):1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999;96(11):6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kashima T, Rao N, Manley JL. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc Natl Acad Sci USA. 2007;104(9):3426–3431. doi: 10.1073/pnas.0700343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnett BG, Muñoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol Cell Biol. 2009;29(5):1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho S, Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24(5):438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh NN, Seo J, Rahn SJ, Singh RN. A multi-exon-skipping detection assay reveals surprising diversity of splice isoforms of spinal muscular atrophy genes. PLoS ONE. 2012;7(11):e49595. doi: 10.1371/journal.pone.0049595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabra M, Texier P, El Maalouf J, Lomonte P. The Tudor protein survival motor neuron (SMN) is a chromatin-binding protein that interacts with methylated lysine 79 of histone H3. J Cell Sci. 2013;126(Pt 16):3664–3677. doi: 10.1242/jcs.126003. [DOI] [PubMed] [Google Scholar]
- 10.Nurputra DK, Lai PS, Harahap NIF, et al. Spinal muscular atrophy: from gene discovery to clinical trials. Ann Hum Genet. 2013;77(5):435–463. doi: 10.1111/ahg.12031. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto T, Sato H, Lai PS, et al. Intragenic mutations in SMN1 may contribute more significantly to clinical severity than SMN2 copy numbers in some spinal muscular atrophy (SMA) patients. Brain Dev. 2013 doi: 10.1016/j.braindev.2013.11.009. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 12.Lefebvre S, Burlet P, Liu Q, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16(3):265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 13.Feldkötter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70(2):358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wirth B, Brichta L, Schrank B, et al. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum Genet. 2006;119(4):422–428. doi: 10.1007/s00439-006-0156-7. [DOI] [PubMed] [Google Scholar]
- 15.Campbell L, Potter A, Ignatius J, Dubowitz V, Davies K. Genomic variation and gene conversion in spinal muscular atrophy: implications for disease process and clinical phenotype. Am J Hum Genet. 1997;61(1):40–50. doi: 10.1086/513886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitali T, Sossi V, Tiziano F, et al. Detection of the survival motor neuron (SMN) genes by FISH: further evidence for a role for SMN2 in the modulation of disease severity in SMA patients. Hum Mol Genet. 1999;8(13):2525–2532. doi: 10.1093/hmg/8.13.2525. [DOI] [PubMed] [Google Scholar]
- 17.Boyer JG, Ferrier A, Kothary R. More than a bystander: the contributions of intrinsic skeletal muscle defects in motor neuron diseases. Front Physiol. 2013;4:356. doi: 10.3389/fphys.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seo J, Howell MD, Singh NN, Singh RN. Spinal muscular atrophy: An update on therapeutic progress. Biochim Biophys Acta. 2013;1832(12):2180–2190. doi: 10.1016/j.bbadis.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang R, So BR, Li P, et al. Structure of a key intermediate of the SMN complex reveals Gemin2’s crucial function in snRNP assembly. Cell. 2011;146(3):384–395. doi: 10.1016/j.cell.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piazzon N, Schlotter F, Lefebvre S, et al. Implication of the SMN complex in the biogenesis and steady state level of the signal recognition particle. Nucleic Acids Res. 2013;41(2):1255–1272. doi: 10.1093/nar/gks1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takaku M, Tsujita T, Horikoshi N, et al. Purification of the human SMN-GEMIN2 complex and assessment of its stimulation of RAD51-mediated DNA recombination reactions. Biochemistry. 2011;50(32):6797–6805. doi: 10.1021/bi200828g. [DOI] [PubMed] [Google Scholar]
- 22.Fallini C, Zhang H, Su Y, et al. The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. J Neurosci. 2011;31(10):3914–3925. doi: 10.1523/JNEUROSCI.3631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez G, Dury AY, Murray LM, et al. A novel function for the survival motoneuron protein as a translational regulator. Hum Mol Genet. 2013;22(4):668–684. doi: 10.1093/hmg/dds474. [DOI] [PubMed] [Google Scholar]
- 24.Bussaglia E, Clermont O, Tizzano E, et al. A frame-shift deletion in the survival motor neuron gene in Spanish spinal muscular atrophy patients. Nat Genet. 1995;11(3):335–337. doi: 10.1038/ng1195-335. [DOI] [PubMed] [Google Scholar]
- 25.Brahe C, Clermont O, Zappata S, Tiziano F, Melki J, Neri G. Frameshift mutation in the survival motor neuron gene in a severe case of SMA type I. Hum Mol Genet. 1996;5(12):1971–1976. doi: 10.1093/hmg/5.12.1971. [DOI] [PubMed] [Google Scholar]
- 26.Parsons DW, McAndrew PE, Monani UR, Mendell JR, Burghes AH, Prior TW. An 11 base pair duplication in exon 6 of the SMN gene produces a type I spinal muscular atrophy (SMA) phenotype: further evidence for SMN as the primary SMA-determining gene. Hum Mol Genet. 1996;5(11):1727–1732. doi: 10.1093/hmg/5.11.1727. [DOI] [PubMed] [Google Scholar]
- 27.Hahnen E, Schönling J, Rudnik-Schöneborn S, Raschke H, Zerres K, Wirth B. Missense mutations in exon 6 of the survival motor neuron gene in patients with spinal muscular atrophy (SMA) Hum Mol Genet. 1997;6(5):821–825. doi: 10.1093/hmg/6.5.821. [DOI] [PubMed] [Google Scholar]
- 28.Talbot K, Ponting CP, Theodosiou AM, et al. Missense mutation clustering in the survival motor neuron gene: a role for a conserved tyrosine and glycine rich region of the protein in RNA metabolism? Hum Mol Genet. 1997;6(3):497–500. doi: 10.1093/hmg/6.3.497. [DOI] [PubMed] [Google Scholar]
- 29.Parsons DW, McAndrew PE, Iannaccone ST, Mendell JR, Burghes AH, Prior TW. Intragenic telSMN mutations: frequency, distribution, evidence of a founder effect, and modification of the spinal muscular atrophy phenotype by cenSMN copy number. Am J Hum Genet. 1998;63(6):1712–1723. doi: 10.1086/302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang CH, Papendick BD, Bruinsma P, Day JK. Identification of a novel missense mutation of the SMN(T) gene in two siblings with spinal muscular atrophy. Neurogenetics. 1998;1(4):273–276. doi: 10.1007/s100480050040. [DOI] [PubMed] [Google Scholar]
- 31.Bühler D, Raker V, Lührmann R, Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum Mol Genet. 1999;8(13):2351–2357. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- 32.Wirth B, Herz M, Wetter A, et al. Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am J Hum Genet. 1999;64(5):1340–1356. doi: 10.1086/302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rochette CF, Suhr LC, Ray PN, et al. Molecular diagnosis of non-deletion SMA patients using quantitative PCR of SMN exon 7. Neurogenetics. 1997;1(2):141–147. doi: 10.1007/s100480050021. [DOI] [PubMed] [Google Scholar]
- 34.Skordis LA, Dunckley MG, Burglen L, et al. Characterisation of novel point mutations in the survival motor neuron gene SMN, in three patients with SMA. Hum Genet. 2001;108(4):356–357. doi: 10.1007/s004390100497. [DOI] [PubMed] [Google Scholar]
- 35.Sossi V, Giuli A, Vitali T, et al. Premature termination mutations in exon 3 of the SMN1 gene are associated with exon skipping and a relatively mild SMA phenotype. Eur J Hum Genet. 2001;9(2):113–120. doi: 10.1038/sj.ejhg.5200599. [DOI] [PubMed] [Google Scholar]
- 36.Tsai CH, Jong YJ, Hu CJ, et al. Molecular analysis of SMN, NAIP and P44 genes of SMA patients and their families. J Neurol Sci. 2001;190(1–2):35–40. doi: 10.1016/s0022-510x(01)00574-3. [DOI] [PubMed] [Google Scholar]
- 37.Martín Y, Valero A, Del Castillo E, Pascual SI, Hernández-Chico C. Genetic study of SMA patients without homozygous SMN1 deletions: identification of compound heterozygotes and characterisation of novel intragenic SMN1 mutations. Hum Genet. 2002;110(3):257–263. doi: 10.1007/s00439-002-0681-y. [DOI] [PubMed] [Google Scholar]
- 38.Cuscó I, López E, Soler-Botija C, Jesús Barceló M, Baiget M, Tizzano EF. A genetic and phenotypic analysis in Spanish spinal muscular atrophy patients with c.399_402del AGAG, the most frequently found subtle mutation in the SMN1 gene. Hum Mutat. 2003;22(2):136–143. doi: 10.1002/humu.10245. [DOI] [PubMed] [Google Scholar]
- 39.Clermont O, Burlet P, Benit P, et al. Molecular analysis of SMA patients without homozygous SMN1 deletions using a new strategy for identification of SMN1 subtle mutations. Hum Mutat. 2004;24(5):417–427. doi: 10.1002/humu.20092. [DOI] [PubMed] [Google Scholar]
- 40.Cuscó I, Barceló MJ, Del Río E, Baiget M, Tizzano EF. Detection of novel mutations in the SMN Tudor domain in type I SMA patients. Neurology. 2004;63(1):146–149. doi: 10.1212/01.wnl.0000132634.48815.13. [DOI] [PubMed] [Google Scholar]
- 41.Sun Y, Grimmler M, Schwarzer V, Schoenen F, Fischer U, Wirth B. Molecular and functional analysis of intragenic SMN1 mutations in patients with spinal muscular atrophy. Hum Mutat. 2005;25(1):64–71. doi: 10.1002/humu.20111. [DOI] [PubMed] [Google Scholar]
- 42.Moutou C, Machev N, Gardes N, Viville S. Case report: birth after preimplantation genetic diagnosis of a subtle mutation in SMN1 gene. Prenat Diagn. 2006;26(11):1037–1041. doi: 10.1002/pd.1551. [DOI] [PubMed] [Google Scholar]
- 43.Kotani T, Sutomo R, Sasongko TH, et al. A novel mutation at the N-terminal of SMN Tudor domain inhibits its interaction with target proteins. J Neurol. 2007;254(5):624–630. doi: 10.1007/s00415-006-0410-x. [DOI] [PubMed] [Google Scholar]
- 44.Zapletalová E, Hedvicáková P, Kozák L, et al. Analysis of point mutations in the SMN1 gene in SMA patients bearing a single SMN1 copy. Neuromuscul Disord. 2007;17(6):476–481. doi: 10.1016/j.nmd.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 45.Brichta L, Garbes L, Jedrzejowska M, et al. Nonsense-mediated messenger RNA decay of survival motor neuron 1 causes spinal muscular atrophy. Hum Genet. 2008;123(2):141–153. doi: 10.1007/s00439-007-0455-7. [DOI] [PubMed] [Google Scholar]
- 46.Eggermann T, Eggermann K, Elbracht M, Zerres K, Rudnik-Schöneborn S. A new splice site mutation in the SMN1 gene causes discrepant results in SMN1 deletion screening approaches. Neuromuscul Disord. 2008;18(2):146–149. doi: 10.1016/j.nmd.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 47.Alías L, Bernal S, Fuentes-Prior P, et al. Mutation update of spinal muscular atrophy in Spain: molecular characterization of 745 unrelated patients and identification of four novel mutations in the SMN1 gene. Hum Genet. 2009;125(1):29–39. doi: 10.1007/s00439-008-0598-1. [DOI] [PubMed] [Google Scholar]
- 48.Prior TW, Krainer AR, Hua Y, et al. A positive modifier of spinal muscular atrophy in the SMN2 gene. Am J Hum Genet. 2009;85(3):408–413. doi: 10.1016/j.ajhg.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernal S, Alías L, Barceló MJ, et al. The c.859G>C variant in the SMN2 gene is associated with types II and III SMA and originates from a common ancestor. J Med Genet. 2010;47(9):640–642. doi: 10.1136/jmg.2010.079004. [DOI] [PubMed] [Google Scholar]
- 50.Sheng-Yuan Z, Xiong F, Chen Y-J, et al. Molecular characterization of SMN copy number derived from carrier screening and from core families with SMA in a Chinese population. Eur J Hum Genet. 2010;18(9):978–984. doi: 10.1038/ejhg.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vezain M, Saugier-Veber P, Goina E, et al. A rare SMN2 variant in a previously unrecognized composite splicing regulatory element induces exon 7 inclusion and reduces the clinical severity of spinal muscular atrophy. Hum Mutat. 2010;31(1):E1110–E1125. doi: 10.1002/humu.21173. [DOI] [PubMed] [Google Scholar]
- 52.Nölle A, Zeug A, Van Bergeijk J, et al. The spinal muscular atrophy disease protein SMN is linked to the Rho-kinase pathway via profilin. Hum Mol Genet. 2011;20(24):4865–4878. doi: 10.1093/hmg/ddr425. [DOI] [PubMed] [Google Scholar]
- 53.Yu-Jin Q, Juan D, Er-zhen L, et al. Subtle mutations in the SMN1 gene in Chinese patients with SMA: p.Arg288Met mutation causing SMN1 transcript exclusion of exon7. BMC Med Genet. 2012;13:86. doi: 10.1186/1471-2350-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kirwin SM, Vinette KMB, Gonzalez IL, Abdulwahed HA, Al-Sannaa N, Funanage VL. A homozygous double mutation in SMN1: a complicated genetic diagnosis of SMA. Mol Genet Genomic Med. 2013;1(2):113–117. doi: 10.1002/mgg3.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ganji H, Nouri N, Salehi M, et al. Detection of intragenic SMN1 mutations in spinal muscular atrophy patients with a single copy of SMN1. J Child Neurol. 2014 doi: 10.1177/0883073814521297. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 56.Mattis VB, Fosso MY, Chang C-W, Lorson CL. Subcutaneous administration of TC007 reduces disease severity in an animal model of SMA. BMC Neurosci. 2009;10:142. doi: 10.1186/1471-2202-10-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finkel RS, Flanigan KM, Wong B, et al. Phase 2a study of ataluren-mediated dystrophin production in patients with nonsense mutation Duchenne muscular dystrophy. PLoS ONE. 2013;8(12):e81302. doi: 10.1371/journal.pone.0081302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mohseni J, Zabidi-Hussin ZAMH, Sasongko TH. Histone deacetylase inhibitors as potential treatment for spinal muscular atrophy. Genet Mol Biol. 2013;36(3):299–307. doi: 10.1590/S1415-47572013000300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowerman M, Beauvais A, Anderson CL, Kothary R. Rho-kinase inactivation prolongs survival of an intermediate SMA mouse model. Hum Mol Genet. 2010;19(8):1468–1478. doi: 10.1093/hmg/ddq021. [DOI] [PubMed] [Google Scholar]
- 60.Bowerman M, Murray LM, Boyer JG, Anderson CL, Kothary R. Fasudil improves survival and promotes skeletal muscle development in a mouse model of spinal muscular atrophy. BMC Med. 2012;10:24. doi: 10.1186/1741-7015-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Z, Kelemen O, Van Santen MA, et al. Synthesis and characterization of pseudocantharidins, novel phosphatase modulators that promote the inclusion of exon 7 into the SMN (survival of motoneuron) pre-mRNA. J Biol Chem. 2011;286(12):10126–10136. doi: 10.1074/jbc.M110.183970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.ClinicalTrialsgov: NCT01302600. www.clinicaltrials.gov/ct2/show/NCT01302600.
- 63.Battle DJ, Kasim M, Yong J, et al. The SMN complex: an assembly machine for RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:313–320. doi: 10.1101/sqb.2006.71.001. [DOI] [PubMed] [Google Scholar]
- 64.Kolb SJ, Battle DJ, Dreyfuss G. Molecular functions of the SMN complex. J Child Neurol. 2007;22(8):990–994. doi: 10.1177/0883073807305666. [DOI] [PubMed] [Google Scholar]
- 65.Strasswimmer J, Lorson CL, Breiding DE, et al. Identification of survival motor neuron as a transcriptional activator-binding protein. Hum Mol Genet. 1999;8(7):1219–1226. doi: 10.1093/hmg/8.7.1219. [DOI] [PubMed] [Google Scholar]
- 66.Campbell L, Hunter KM, Mohaghegh P, Tinsley JM, Brasch MA, Davies KE. Direct interaction of Smn with dp103, a putative RNA helicase: a role for Smn in transcription regulation? Hum Mol Genet. 2000;9(7):1093–1100. doi: 10.1093/hmg/9.7.1093. [DOI] [PubMed] [Google Scholar]
- 67.Bowerman M, Shafey D, Kothary R. Smn depletion alters profilin II expression and leads to upregulation of the RhoA/ROCK pathway and defects in neuronal integrity. J Mol Neurosci. 2007;32(2):120–131. doi: 10.1007/s12031-007-0024-5. [DOI] [PubMed] [Google Scholar]
- 68.Zou T, Yang X, Pan D, Huang J, Sahin M, Zhou J. SMN deficiency reduces cellular ability to form stress granules, sensitizing cells to stress. Cell Mol Neurobiol. 2011;31(4):541–550. doi: 10.1007/s10571-011-9647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rossoll W, Kröning A-K, Ohndorf U-M, Steegborn C, Jablonka S, Sendtner M. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum Mol Genet. 2002;11(1):93–105. doi: 10.1093/hmg/11.1.93. [DOI] [PubMed] [Google Scholar]
- 70.Peter CJ, Evans M, Thayanithy V, et al. The COPI vesicle complex binds and moves with survival motor neuron within axons. Hum Mol Genet. 2011;20(9):1701–1711. doi: 10.1093/hmg/ddr046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Akten B, Kye MJ, Hao LT, et al. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc Natl Acad Sci USA. 2011;108(25):10337–10342. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fallini C, Bassell GJ, Rossoll W. Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res. 2012;1462:81–92. doi: 10.1016/j.brainres.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grimmler M, Bauer L, Nousiainen M, Körner R, Meister G, Fischer U. Phosphorylation regulates the activity of the SMN complex during assembly of spliceosomal U snRNPs. EMBO Rep. 2005;6(1):70–76. doi: 10.1038/sj.embor.7400301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Renvoisé B, Quérol G, Verrier ER, Burlet P, Lefebvre S. A role for protein phosphatase PP1? in SMN complex formation and subnuclear localization to Cajal bodies. J Cell Sci. 2012;125(Pt 12):2862–2874. doi: 10.1242/jcs.096255. [DOI] [PubMed] [Google Scholar]
- 75.Wu C-Y, Curtis A, Choi YS, et al. Identification of the phosphorylation sites in the survival motor neuron protein by protein kinase A. Biochim Biophys Acta. 2011;1814(9):1134–1139. doi: 10.1016/j.bbapap.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Makhortova NR, Hayhurst M, Cerqueira A, et al. A screen for regulators of survival of motor neuron protein levels. Nat Chem Biol. 2011;7(8):544–552. doi: 10.1038/nchembio.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Setola V, Terao M, Locatelli D, Bassanini S, Garattini E, Battaglia G. Axonal-SMN (a-SMN), a protein isoform of the survival motor neuron gene, is specifically involved in axonogenesis. Proc Natl Acad Sci USA. 2007;104(6):1959–1964. doi: 10.1073/pnas.0610660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gunadi, Sasongko TH, Yusoff S, et al. Hypomutability at the polyadenine tract in SMN intron 3 shows the invariability of the a-SMN protein structure. Ann Hum Genet. 2008;72(Pt 2):288–291. doi: 10.1111/j.1469-1809.2007.00409.x. [DOI] [PubMed] [Google Scholar]
- 79.Locatelli D, Terao M, Fratelli M, et al. Human axonal survival of motor neuron (a-SMN) protein stimulates axon growth, cell motility, C-C motif ligand 2 (CCL2), and insulin-like growth factor-1 (IGF1) production. J Biol Chem. 2012;287(31):25782–25794. doi: 10.1074/jbc.M112.362830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Le TT, McGovern VL, Alwine IE, et al. Temporal requirement for high SMN expression in SMA mice. Hum Mol Genet. 2011;20(18):3578–3591. doi: 10.1093/hmg/ddr275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lutz CM, Kariya S, Patruni S, et al. Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J Clin Invest. 2011;121(8):3029–3041. doi: 10.1172/JCI57291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hao LT, Duy PQ, Jontes JD, Wolman M, Granato M, Beattie CE. Temporal requirement for SMN in motoneuron development. Hum Mol Genet. 2013;22(13):2612–2625. doi: 10.1093/hmg/ddt110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kariya S, Obis T, Garone C, et al. Requirement of enhanced Survival Motoneuron protein imposed during neuromuscular junction maturation. J Clin Invest. 2014;124(2):785–800. doi: 10.1172/JCI72017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shababi M, Lorson CL, Rudnik-Schöneborn SS. Spinal muscular atrophy: a motor neuron disorder or a multi-organ disease? J Anat. 2014;224(1):15–28. doi: 10.1111/joa.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30(4):377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 86.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34(4):460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 87.Singh NN, Androphy EJ, Singh RN. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem Biophys Res Commun. 2004;315(2):381–388. doi: 10.1016/j.bbrc.2004.01.067. [DOI] [PubMed] [Google Scholar]
- 88.Singh NN, Singh RN. Alternative splicing in spinal muscular atrophy underscores the role of an intron definition model. RNA Biol. 2011;8(4):600–606. doi: 10.4161/rna.8.4.16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singh NN, Androphy EJ, Singh RN. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10(8):1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singh NN, Seo J, Ottesen EW, Shishimorova M, Bhattacharya D, Singh RN. TIA1 prevents skipping of a critical exon associated with spinal muscular atrophy. Mol Cell Biol. 2011;31(5):935–954. doi: 10.1128/MCB.00945-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singh NN, Singh RN, Androphy EJ. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007;35(2):371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Singh NN, Lawler MN, Ottesen EW, Upreti D, Kaczynski JR, Singh RN. An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy. Nucleic Acids Res. 2013;41(17):8144–8165. doi: 10.1093/nar/gkt609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5(4):e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Singh NK, Singh NN, Androphy EJ, Singh RN. Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol. 2006;26(4):1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6(3):341–350. doi: 10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet. 2008;82(4):834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Singh NN, Hollinger K, Bhattacharya D, Singh RN. An antisense microwalk reveals critical role of an intronic position linked to a unique long-distance interaction in pre-mRNA splicing. RNA. 2010;16(6):1167–1181. doi: 10.1261/rna.2154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Klar J, Sobol M, Melberg A, et al. Welander distal myopathy caused by an ancient founder mutation in TIA1 associated with perturbed splicing. Hum Mutat. 2013;34(4):572–577. doi: 10.1002/humu.22282. [DOI] [PubMed] [Google Scholar]
- 99.Keil J, Seo J, Howell MD, Hsu W, Singh RN, DiDonato CJ. A short antisense oligonucleotide ameliorates symptoms of severe mouse models of spinal muscular atrophy. Mol Ther Nucleic Acids. 2014 doi: 10.1038/mtna.2014.23. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee RG, Crosby J, Baker BF, Graham MJ, Crooke RM. Antisense technology: an emerging platform for cardiovascular disease therapeutics. J Cardiovasc Transl Res. 2013;6(6):969–980. doi: 10.1007/s12265-013-9495-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roberts TC, Wood MJA. Therapeutic targeting of non-coding RNAs. Essays Biochem. 2013;54:127–145. doi: 10.1042/bse0540127. [DOI] [PubMed] [Google Scholar]
- 102.Sivanesan S, Howell MD, DiDonato CJ, Singh RN. Antisense oligonucleotide mediated therapy of spinal muscular atrophy. Translat Neurosci. 2013;4(1):1–7. doi: 10.2478/s13380-013-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Porensky PN, Burghes AHM. Antisense oligonucleotides for the treatment of spinal muscular atrophy. Hum Gene Ther. 2013;24(5):489–498. doi: 10.1089/hum.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Buratti E, Baralle M, Baralle FE. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34(12):3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hua Y, Sahashi K, Rigo F, et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478(7367):123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Porensky PN, Mitrpant C, McGovern VL, et al. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum Mol Genet. 2012;21(7):1625–1638. doi: 10.1093/hmg/ddr600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou H, Janghra N, Mitrpant C, et al. A novel morpholino oligomer targeting ISS-N1 improves rescue of severe spinal muscular atrophy transgenic mice. Hum Gene Ther. 2013;24(3):331–342. doi: 10.1089/hum.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mitrpant C, Porensky P, Zhou H, et al. Improved antisense oligonucleotide design to suppress aberrant SMN2 gene transcript processing: towards a treatment for spinal muscular atrophy. PLoS ONE. 2013;8(4):e62114. doi: 10.1371/journal.pone.0062114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.ClinicalTrialsgov: NCT01839656. www.clinicaltrials.gov/show/NCT01839656.
- 110.ClinicalTrialsgov: NCT01494701. www.clinicaltrials.gov/ct2/show/NCT01494701.
- 111.Friedman Y, Balaga O, Linial M. Working together: combinatorial regulation by microRNAs. Adv Exp Med Biol. 2013;774:317–337. doi: 10.1007/978-94-007-5590-1_16. [DOI] [PubMed] [Google Scholar]
- 112.Brosseau J-P, Lucier J-F, Lamarche A-A, et al. Redirecting splicing with bifunctional oligonucleotides. Nucl Acids Res. 2014;42(6):e40–e40. doi: 10.1093/nar/gkt1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Obad S, Dos Santos CO, Petri A, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43(4):371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Echaniz-Laguna A, Miniou P, Bartholdi D, Melki J. The promoters of the survival motor neuron gene (SMN) and its copy (SMNc) share common regulatory elements. Am J Hum Genet. 1999;64(5):1365–1370. doi: 10.1086/302372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Monani UR, Lorson CL, Parsons DW, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8(7):1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 116.Germain-Desprez D, Brun T, Rochette C, Semionov A, Rouget R, Simard LR. The SMN genes are subject to transcriptional regulation during cellular differentiation. Gene. 2001;279(2):109–117. doi: 10.1016/s0378-1119(01)00758-2. [DOI] [PubMed] [Google Scholar]
- 117.Boda B, Mas C, Giudicelli C, et al. Survival motor neuron SMN1 and SMN2 gene promoters: identical sequences and differential expression in neurons and non-neuronal cells. Eur J Hum Genet. 2004;12(9):729–737. doi: 10.1038/sj.ejhg.5201217. [DOI] [PubMed] [Google Scholar]
- 118.Branchu J, Biondi O, Chali F, et al. Shift from extracellular signal-regulated kinase to AKT/cAMP response element-binding protein pathway increases survival-motor-neuron expression in spinal-muscular-atrophy-like mice and patient cells. J Neurosci. 2013;33(10):4280–4294. doi: 10.1523/JNEUROSCI.2728-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Farooq F, Molina FA, Hadwen J, et al. Prolactin increases SMN expression and survival in a mouse model of severe spinal muscular atrophy via the STAT5 pathway. J Clin Invest. 2011;121(8):3042–3050. doi: 10.1172/JCI46276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Biondi O, Branchu J, Sanchez G, et al. In vivo NMDA receptor activation accelerates motor unit maturation, protects spinal motor neurons, and enhances SMN2 gene expression in severe spinal muscular atrophy mice. J Neurosci. 2010;30(34):11288–11299. doi: 10.1523/JNEUROSCI.1764-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kolb EA, Gorlick R, Houghton PJ, et al. Initial testing (stage 1) of AZD6244 (ARRY-142886) by the Pediatric Preclinical Testing Program. Pediatr Blood Cancer. 2010;55(4):668–677. doi: 10.1002/pbc.22576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Majumder S, Varadharaj S, Ghoshal K, Monani U, Burghes AHM, Jacob ST. Identification of a novel cyclic AMP-response element (CRE-II) and the role of CREB-1 in the cAMP-induced expression of the survival motor neuron (SMN) gene. J Biol Chem. 2004;279(15):14803–14811. doi: 10.1074/jbc.M308225200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 123.Besnard A, Galan-Rodriguez B, Vanhoutte P, Caboche J. Elk-1 a transcription factor with multiple facets in the brain. Front Neurosci. 2011;5:35. doi: 10.3389/fnins.2011.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou Z, Qiu J, Liu W, et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell. 2012;47(3):422–433. doi: 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Swoboda KJ, Scott CB, Reyna SP, et al. Phase II open label study of valproic acid in spinal muscular atrophy. PLoS ONE. 2009;4(5):e5268. doi: 10.1371/journal.pone.0005268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Swoboda KJ, Scott CB, Crawford TO, et al. SMA CARNIVAL trial part I: double-blind, randomized, placebo-controlled trial of L-carnitine and valproic acid in spinal muscular atrophy. PLoS ONE. 2010;5(8):e12140. doi: 10.1371/journal.pone.0012140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen T-H, Chang J-G, Yang Y-H, et al. Randomized, double-blind, placebo-controlled trial of hydroxyurea in spinal muscular atrophy. Neurology. 2010;75(24):2190–2197. doi: 10.1212/WNL.0b013e3182020332. [DOI] [PubMed] [Google Scholar]
- 128.Kissel JT, Scott CB, Reyna SP, et al. SMA carnival trial part II: a prospective, single-armed trial of L-carnitine and valproic acid in ambulatory children with spinal muscular atrophy. PLoS ONE. 2011;6(7):e21296. doi: 10.1371/journal.pone.0021296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Darbar IA, Plaggert PG, Resende MBD, Zanoteli E, Reed UC. Evaluation of muscle strength and motor abilities in children with type II and III spinal muscle atrophy treated with valproic acid. BMC Neurol. 2011;11:36. doi: 10.1186/1471-2377-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu H, Rodgers ND, Jiao X, Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21(17):4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jarecki J, Chen X, Bernardino A, et al. Diverse small-molecule modulators of SMN expression found by high-throughput compound screening: early leads towards a therapeutic for spinal muscular atrophy. Hum Mol Genet. 2005;14(14):2003–2018. doi: 10.1093/hmg/ddi205. [DOI] [PubMed] [Google Scholar]
- 132.Van Meerbeke JP, Gibbs RM, Plasterer HL, et al. The DcpS inhibitor RG3039 improves motor function in SMA mice. Hum Mol Genet. 2013;22(20):4074–4083. doi: 10.1093/hmg/ddt257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gogliotti RG, Cardona H, Singh J, et al. The DcpS inhibitor RG3039 improves survival, function and motor unit pathologies in two SMA mouse models. Hum Mol Genet. 2013;22(20):4084–4101. doi: 10.1093/hmg/ddt258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Azzouz M, Le T, Ralph GS, et al. Lentivector-mediated SMN replacement in a mouse model of spinal muscular atrophy. J Clin Invest. 2004;114(12):1726–1731. doi: 10.1172/JCI22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dayton RD, Wang DB, Klein RL. The advent of AAV9 expands applications for brain and spinal cord gene delivery. Expert Opin Biol Ther. 2012;12(6):757–766. doi: 10.1517/14712598.2012.681463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mendell JR, Rodino-Klapac L, Sahenk Z, et al. Gene therapy for muscular dystrophy: lessons learned and path forward. Neurosci Lett. 2012;527(2):90–99. doi: 10.1016/j.neulet.2012.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Passini MA, Bu J, Roskelley EM, et al. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J Clin Invest. 2010;120(4):1253–1264. doi: 10.1172/JCI41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Foust KD, Wang X, McGovern VL, et al. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol. 2010;28(3):271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 139.Bevan AK, Hutchinson KR, Foust KD, et al. Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum Mol Genet. 2010;19(20):3895–3905. doi: 10.1093/hmg/ddq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shababi M, Habibi J, Ma L, Glascock JJ, Sowers JR, Lorson CL. Partial restoration of cardio-vascular defects in a rescued severe model of spinal muscular atrophy. J Mol Cell Cardiol. 2012;52(5):1074–1082. doi: 10.1016/j.yjmcc.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dominguez E, Marais T, Chatauret N, et al. Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice. Hum Mol Genet. 2011;20(4):681–693. doi: 10.1093/hmg/ddq514. [DOI] [PubMed] [Google Scholar]
- 142.Valori CF, Ning K, Wyles M, et al. Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci Transl Med. 2010;2(35):35ra42. doi: 10.1126/scitranslmed.3000830. [DOI] [PubMed] [Google Scholar]
- 143.Glascock JJ, Shababi M, Wetz MJ, Krogman MM, Lorson CL. Direct central nervous system delivery provides enhanced protection following vector mediated gene replacement in a severe model of spinal muscular atrophy. Biochem Biophys Res Commun. 2012;417(1):376–381. doi: 10.1016/j.bbrc.2011.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Benkhelifa-Ziyyat S, Besse A, Roda M, et al. Intramuscular scAAV9-SMN injection mediates widespread gene delivery to the spinal cord and decreases disease severity in SMA mice. Mol Ther. 2013;21(2):282–290. doi: 10.1038/mt.2012.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Families of SMA. US FDA approval to Nationwide Children’s Hosptial for Phase I clinical trial of systemic AAV9-delivered SMN gene for spinal muscular atrophy. www.fsma.org/LatestNews/index.cfm?ID=8172&TYPE=1150.
- 146.Oprea GE, Kröber S, McWhorter ML, et al. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320(5875):524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Amara A, Adala L, Ben Charfeddine I, et al. Correlation of SMN2, NAIP, p44, H4F5 and Occludin genes copy number with spinal muscular atrophy phenotype in Tunisian patients. Eur J Paediatr Neurol. 2012;16(2):167–174. doi: 10.1016/j.ejpn.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 148.Zhang Z, Pinto AM, Wan L, et al. Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc Natl Acad Sci USA. 2013;110(48):19348–19353. doi: 10.1073/pnas.1319280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ahmad S, Wang Y, Shaik GM, Burghes AH, Gangwani L. The zinc finger protein ZPR1 is a potential modifier of spinal muscular atrophy. Hum Mol Genet. 2012;21(12):2745–2758. doi: 10.1093/hmg/dds102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Perrin S. Preclinical research: Make mouse studies work. Nature. 2014;507(7493):423–425. doi: 10.1038/507423a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.