Table 1. Data-collection statistics and structure-refinement parameters.
Values in parentheses are for the highest resolution shell.
| LLT1_mono | LLT1_D1 | LLT1_D2 | LLT1_glyco | |
|---|---|---|---|---|
| PDB code | 4qkg | 4qkh | 4qki | 4qkj |
| pH of crystallization condition | 3.5 | 7.0 | 7.0 | 4.2 |
| Glycosylation | GlcNAc | GlcNAc | GlcNAc | GlcNAc2Man5 |
| Oligomer | Monomer | Dimer | Dimer | Hexamer |
| Data-collection statistics | ||||
| Resolution range () | 50.01.95 (1.981.95) | 47.31.8 (1.841.80) | 43.71.8 (1.841.80) | 39.02.7 (2.762.70) |
| Space group | P3221 | P212121 | P212121 | P6322 |
| Unit-cell parameters () | a = b = 47.3, c = 106.1 | a = 50.9, b = 57.8, c = 82.3 | a = 51.3, b = 54.1, c = 74.2 | a = b = 70.1, c = 101.7 |
| Radiation source | I02, DLS | I04-1, DLS | I04-1, DLS | BM 14.1, BESSY II |
| Detector | ADSC Q315 CCD | PILATUS 2M | PILATUS 2M | MAR Mosaic 225 CCD |
| Data-processing software | HKL-2000 [DENZO, SCALEPACK] | MOSFLM, AIMLESS | MOSFLM, AIMLESS | HKL-2000 [DENZO, SCALEPACK], anisotropy server |
| Wavelength () | 0.9796 | 0.9200 | 0.9200 | 0.9184 |
| No. of images | 220 | 900 | 1800 | 80 [images 150, 330360] |
| Crystal-to-detector distance (mm) | 256.1 | 325.2 | 251.9 | 288.1 |
| Exposure time per image (s) | 1 | 0.2 | 0.2 | 2 |
| Oscillation width () | 0.5 | 0.2 | 0.2 | 0.5 |
| No. of observations | 66670 (3340) | 175131 (5869) | 113353 (6521) | 478731 (1201) |
| No. of unique reflections | 10555 (506) | 21104 (1087) | 18996 (1050) | 4431 (273) |
| Data completeness (%) | 99.7 (100) | 91.8 (81.0) | 96.7 (91.6) | 99.5 (99.3) |
| Average multiplicity | 6.3 (6.6) | 8.3 (5.4) | 6.0 (6.2) | 4.3 (4.4) |
| Mosaicity () | 0.30.7 | 1.8 | 1.3 | 0.81.4 |
| I/(I) | 21.2 (2.9) | 11.3 (3.0) | 11.8 (3.7) | 16.2 (2.0) |
| Solvent content (%) | 44 | 36 | 25 | 46 |
| Matthews coefficient (3Da1) | 2.18 | 1.93 | 1.64 | 2.30 |
| B factor from Wilson plot (2) | 34.0 | 20.7 | 17.7 | 66.1 |
| R merge † | 0.074 (0.599) | 0.093 (0.462) | 0.082 (0.404) | 0.096 (0.757) |
| R p.i.m. ‡ | 0.032 (0.251) | 0.045 (0.309) | 0.053 (0.257) | 0.031 (0.171) |
| Structure parameters | ||||
| R work § (%) | 19.4 | 17.5 | 17.8 | 22.1 |
| R free § (%) | 24.7 | 25.0 | 24.4 | 29.8 |
| R all § (%) | 20.1 | 18.0 | 18.3 | 23.1 |
| Average B factor (2) | 39.8 | 27.6 | 20.9 | 40.1 |
| R.m.s.d. from ideal bond lengths () | 0.016 | 0.016 | 0.016 | 0.017 |
| R.m.s.d. from ideal bond angles () | 1.9 | 1.7 | 1.8 | 2.0 |
| No. of monomers per asymmetric unit | 1 | 2 | 2 | 1 |
| Amino-acid residues located | A72A146, A161A193 | A70A192, B72B192 | A73A194, B73B192 | A74A188 |
| Asn residues with located GlcNAc | A95 | A95, A147, B95, B147 | A95, A147, B95, B147 | A95 |
| No. of water molecules | 42 | 215 | 167 | 15 |
| Other localized moieties | 1 SO4 2 | |||
| Ramachandran statistics: residues in favoured region (%) | 95 | 98 | 98 | 87 |
R
merge = 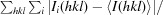
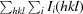 (Diederichs Karplus, 1997 ▶), where Ii(hkl) and I(hkl) are the observed individual and mean intensities of a reflection with indices hkl, respectively,
(Diederichs Karplus, 1997 ▶), where Ii(hkl) and I(hkl) are the observed individual and mean intensities of a reflection with indices hkl, respectively, 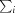 is the sum over the individual measurements of a reflection with indices hkl and
is the sum over the individual measurements of a reflection with indices hkl and 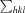 is the sum over all reflections.
is the sum over all reflections.
R
p.i.m. = 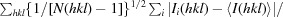
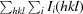 (Weiss, 2001 ▶), where N(hkl) is the redundancy of the reflection with indices hkl.
(Weiss, 2001 ▶), where N(hkl) is the redundancy of the reflection with indices hkl.
R
work = 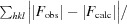
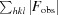 , where F
obs and F
calc are the observed and calculated structure-factor amplitudes for the reflection with indices hkl for the working set of reflections. R
free is the same as R
work but is calculated for 56% of the data omitted from refinement. R
all sums over all reflections.
, where F
obs and F
calc are the observed and calculated structure-factor amplitudes for the reflection with indices hkl for the working set of reflections. R
free is the same as R
work but is calculated for 56% of the data omitted from refinement. R
all sums over all reflections.
