Abstract
The GSTT1 gene encodes a key enzyme involved in the metabolism of xenobiotics and its polymorphisms have been associated with individual susceptibility to various malignancies. Numerous molecular epidemiological studies have been performed to investigate the association between GSTT1 gene polymorphisms and lung cancer susceptibility; however, the results of previous studies were inconsistent. Therefore, the aim of the present study was to conduct a meta-analysis in order to derive a more precise estimation of the association in the East Asian populations. The meta-analysis included 7,415 lung cancer cases and 6,084 controls from 26 published studies in East Asia, which were selected from the PubMed and China National Knowledge Infrastructure databases, up to March 20, 2014. Using crude odds ratios (ORs) with 95% confidence intervals (CIs), a statistically significant association was identified between the GSTT1 null genotype and lung cancer in the East Asian populations (OR=1.17; 95% CI, 1.09–1.25; Pheterogeneity=0.003). Furthermore, subgroup analyses revealed that the lung cancer risk in smokers carrying the GSTT1 null genotype was significantly increased compared with non-smokers (OR=1.71; 95% CI, 1.04–2.81; Pheterogeneity=0.002). Thus, the GSTT1 null genotype may increase the risk of lung cancer among the East Asian populations.
Keywords: meta-analysis, glutathione S-transferase θ1, polymorphism, lung cancer
Introduction
Lung cancer is the most common type of cancer and the leading cause of cancer-associated mortality in the world, becoming a major public health problem globally. Lung cancer accounts for 13% (1.6 million) of the total number of cancer cases and 18% (1.4 million) of mortalities in 2008 (1). In recent years, the incidence of lung cancer in Asia has increased rapidly; however, the etiological factors of the disease remain unclear. Lung cancer is known to be a complex disease; however, tobacco smoking, family history, diet and susceptible gene mutations appear to be involved in its development (1,2).
Glutathione S-transferases (GSTs) are phase II enzymes that are key in the detoxification of numerous carcinogens (3). GSTs mediate the conjugation of electrophilic compounds to glutathione, resulting in detoxification of specific environmental carcinogens and pesticides and the inactivation of polycyclic aromatic hydrocarbons, facilitating their excretion from the body (4,5). GSTs are classified into at least four genetically distinct groups: α, μ, π and θ. Theoretically, individuals lacking a specific GST enzyme may be at a particularly high risk of developing cancer, if exposed to certain genotoxicants (4,6). Among the potential mutations, the most widely known are the deletions of the GST θ1 (GSTT1) or GST μ1 (GSTM1) genes (null variants), which result in no enzymatic functional activity (7,8). Deficiency in GSTT1 isoenzyme activity may predispose individuals to the effects of electrophilic carcinogens. Previous studies have proposed that GSTT1 deficiencies may be associated with an increased susceptibility to lung cancer (9–12); however, other studies contradict this proposal (13–15). Therefore, numerous molecular epidemiological studies have been performed to investigate the potential association between the GSTT1 gene polymorphism and lung cancer susceptibility, particularly in East Asian populations (15–17); however, the conclusions are inconsistent and occasionally contradictory. A previous study has indicated that the frequency of the GSTT1 null genotype is higher in Asia compared with other populations (18). Thus, the present study performed a meta-analysis, including 26 eligible case-control or prospective studies (15–17,19–41), aiming to investigate the effect of the GSTT1 polymorphism on the risk of lung cancer in the East Asian populations.
Materials and methods
Search strategy
Eligible studies were identified by performing a literature search on the PubMed and China National Knowledge Infrastructure (CNKI) databases (from inception to March 20, 2014) using the following keywords: ‘Glutathione S-transferase T1’, ‘GSTT1’, ‘polymorphism’, ‘lung cancer’ and the combined phrases. In addition, the references of all the relevant articles and reviews were searched for additional eligible studies. Furthermore, the full text of each potentially relevant paper was scrutinized to ensure that the following inclusion criteria were met: i) The studies had an observational (case-control or prospective) study design; ii) the authors offered sufficient data for estimating odds ratios (ORs) and their 95% confidence intervals (CIs); and, iii) the patients and controls of all the studies were East Asian (including Chinese, Japanese, South Korean, Mongolian and North Korean). In cases where multiple studies reporting on the same population data met the inclusion criteria, the study with the largest sample size was selected for subsequent analysis.
Data extraction
In order to minimize bias, the following data were extracted from all the eligible studies by two researchers independently: First author’s surname, year of publication, country, source of the controls (population- or hospital-based studies), histological type of cancer (adenocarcinoma, squamous cell carcinoma or small-cell carcinoma), smoking status and the different genotypes in the cases and controls. In accordance with the definition used in the majority of previous studies, carriers with at least one GSTT1 allele were defined as the ‘present’ genotype group, whereas individuals carrying no GSTT1 alleles were classified as the ‘null’ genotype group.
Quality assessment
The quality of the studies included in the present analysis were assessed using an adapted 10-point Newcastle-Ottawa assessment scale (NOS) (42). The quality of each study was assessed by two independent reviewers based on three broad factors: Selection (maximum score, 4), comparability (maximum score, 2) and exposure (maximum score, 4). Thus, a total quality score ranging between 0 (lowest score) and 10 (highest score) was obtained by adding all the scores. A total score of seven or greater indicated that the study was of high-quality.
Statistical analysis
The association between lung cancer risk and the GSTT1 polymorphism was estimated for each study using crude ORs with 95% CIs. The pooled ORs were evaluated for null vs. present genotypes and a χ2-based Q-statistical test was performed to assess the heterogeneity between the studies (43). P<0.05 was considered to indicate a statistically significant heterogeneity. In the case of significant heterogeneity, a random-effect model, as described by DerSimonian and Laird (44), was used to calculate pooled estimates. Otherwise, a fixed-effect model was used, as described by the Mantel-Haenszel method (45). These two models provided similar results when heterogeneity between the studies was absent. The potential publication bias was evaluated using the funnel plot and the linear regression asymmetry test, as previously described by Egger et al (46). P<0.05 was considered to indicate a statistical significant difference. All the statistical analyses were performed using the Statistical Analysis System (version 9.1.3; SAS Institute, Cary, NC, USA) and Review Manager software (version 5.2; The Cochrane Collaboration, Oxford, UK), with two-sided P-values.
Results
Eligible studies
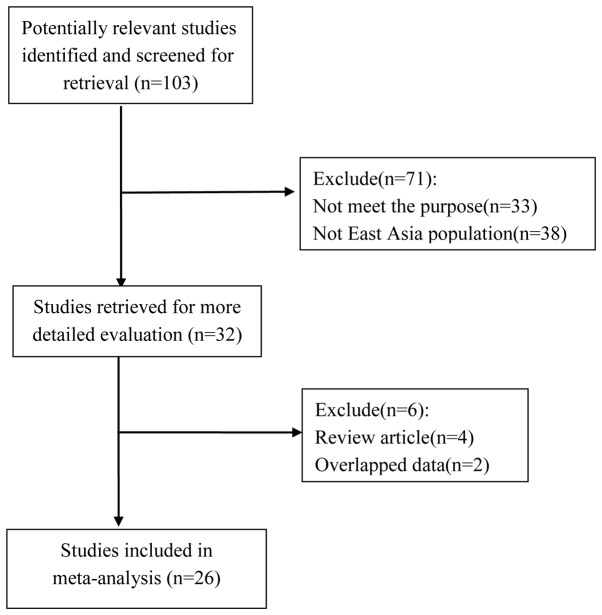
Fig. 1 indicates the process of study selection and exclusion. A total of 103 abstracts were identified in PubMed and CNKI using the aforementioned key words. Following careful review of the titles and abstracts, 32 relevant studies describing the association between GSTT1 polymorphisms and lung cancer in the East Asian populations were selected. However, after obtaining and reading the full articles, four studies were excluded since they were review articles and two studies were excluded since they presented no data of interest or only raw data. Thus, a total of 26 eligible studies, including 7,415 lung cancer cases and 6,084 controls, were selected for inclusion in the present meta-analysis. The major characteristics of these studies are presented in Table I. The sample size range was 107–5,632 samples, while 10 studies used hospital-based control sources, 16 studies were hospital-based and two studies did not specify. In addition, sub-analyses by histological type (adenocarcinoma, 7; squamous cell carcinoma, 7; small-cell carcinoma, 3) and smoking state (smokers, 8; non-smokers, 8) were conducted. The methodological quality of the studies included is presented in Table I and the Newcastle-Ottawa assessment scale score range was 4–9, with a mean score of 6.7.
Figure 1.
Flow chart of the selection of publications included in the current meta-analysis.
Table I.
Characteristics of studies included in the present meta-analysis.
| Genotypic distribution | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
| Case | Control | Newcastle-Ottawa scale | |||||||||
|
|
|
|
|||||||||
| Reference | Year | Country | Controls | Null | Present | Null | Present | Selectiona | Comparabilityb | Assessmenta | Total |
| Piao et al (17) | 2013 | Korean | HB | 2070 | 1863 | 858 | 841 | 3 | 2 | 2 | 7 |
| Wang et al (27) | 2012 | China | PB | 90 | 119 | 100 | 156 | 3 | 2 | 2 | 7 |
| Liang et al (41) | 2012 | China | HB | 45 | 23 | 34 | 36 | 2 | 0 | 2 | 4 |
| Kiyohara et al (15) | 2012 | Japan | PB | 217 | 245 | 164 | 215 | 3 | 2 | 4 | 9 |
| Liu (34) | 2011 | China | HB | 57 | 43 | 56 | 79 | 3 | 0 | 3 | 6 |
| Tamaki et al (26) | 2011 | Japan | PB | 97 | 95 | 104 | 99 | 3 | 1 | 3 | 7 |
| Fang (33) | 2011 | China | HB | 45 | 23 | 34 | 36 | 3 | 0 | 2 | 5 |
| Du (31) | 2011 | China | HB | 57 | 68 | 56 | 69 | 3 | 1 | 2 | 6 |
| Bai (29) | 2011 | China | HB | 56 | 72 | 96 | 118 | 3 | 1 | 2 | 6 |
| Fan et al (39) | 2010 | China | HB | 38 | 20 | 29 | 31 | 3 | 2 | 2 | 7 |
| Qi et al (37) | 2008 | China | HB | 17 | 36 | 27 | 45 | 3 | 2 | 2 | 7 |
| Yang et al (25) | 2007 | Korea | HB | 168 | 148 | 166 | 179 | 3 | 2 | 2 | 7 |
| Li (30) | 2007 | China | HB | 17 | 25 | 48 | 55 | 3 | 1 | 2 | 6 |
| Lee et al (23) | 2006 | Korea | HB | 80 | 89 | 107 | 89 | 2 | 2 | 2 | 6 |
| He and Tang (35) | 2006 | China | PB | 33 | 28 | 29 | 17 | 4 | 0 | 3 | 7 |
| Chen et al (20) | 2006 | China | PB | 59 | 38 | 85 | 112 | 3 | 0 | 2 | 5 |
| Yuan et al (28) | 2005 | China | HB | 82 | 68 | 58 | 94 | 3 | 1 | 2 | 6 |
| Cao et al (40) | 2004 | China | HB | 69 | 35 | 87 | 118 | 2 | 1 | 2 | 5 |
| Chan-Yeung et al (19) | 2004 | China | PB | 143 | 86 | 102 | 95 | 4 | 2 | 2 | 8 |
| Wang et al (5) | 2003 | China | NR | 53 | 59 | 54 | 65 | 2 | 1 | 3 | 6 |
| Wang (32) | 2003 | China | HB | 44 | 33 | 54 | 53 | 3 | 0 | 3 | 6 |
| Zhang (38) | 2002 | China | PB | 74 | 87 | 72 | 93 | 3 | 2 | 3 | 8 |
| London et al (24) | 2000 | China | PB | 134 | 98 | 426 | 284 | 4 | 2 | 3 | 9 |
| Lan et al (22) | 2000 | China | PB | 73 | 49 | 64 | 58 | 3 | 2 | 3 | 8 |
| Kiyohara et al (22) | 2000 | Japan | NR | 47 | 14 | 39 | 12 | 3 | 2 | 4 | 9 |
| Lan et al (37) | 1991 | China | PB | 52 | 34 | 52 | 34 | 4 | 2 | 2 | 8 |
Maximum score, 4;
maximum score, 2.
HB, hospital-based; PB, population-based.
Meta-analysis
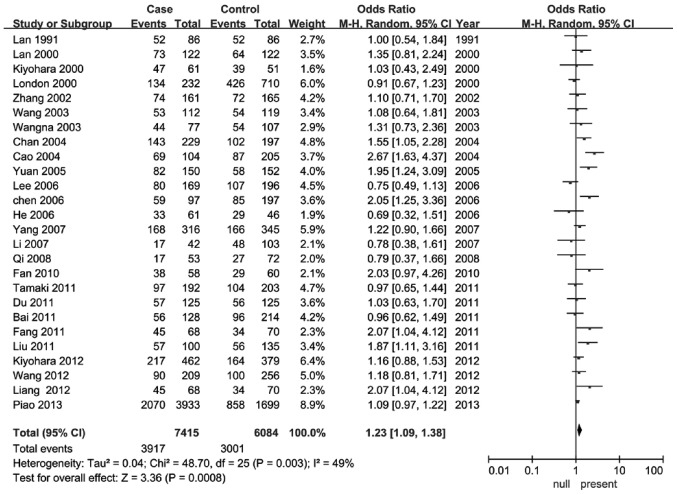
As indicated in Table II, the pooled ORs were performed for the GSTT1 null vs. present genotype individuals. The results indicated that individuals with the GSTT1 null genotype were significantly associated with an increased risk of developing lung cancer compared with those carrying the GSTT1 present genotype in the East Asian populations (OR, 1.23; 95% CI, 1.09–1.38; Pheterogeneity=0.003; Fig. 2). When stratified by smoking status, a statistically increased lung cancer risk was identified in smokers (OR, 1.71; 95% CI=1.04–2.81; Pheterogeneity=0.002). However, no statistically significant association was identified in non-smokers (OR=1.13; 95% CI, 0.87–1.46; Pheterogeneity=0.42). Furthermore, in subgroup analysis by histological type, no statistically significant association was identified for all the stratified analyses. The major results of the meta-analysis and the heterogeneity test are listed in Table II.
Table II.
Main results of pooled ORs in the meta-analysis.
| Null versus present | ||||
|---|---|---|---|---|
|
|
||||
| Studies, n | OR | 95% CI | P-valuea | |
| Total | 26 | 1.23 | 1.09–1.38 | 0.003 |
| Histological type | ||||
| SCLC | 3 | 1.17 | 0.69–1.98 | 0.97 |
| SCC | 7 | 1.33 | 0.83–2.15 | 0.001 |
| AC | 7 | 1.24 | 0.98–1.57 | 0.10 |
| Smoking status | ||||
| Smoker | 8 | 1.71 | 1.04–2.81 | 0.002 |
| Non-smoker | 8 | 1.13 | 0.87–1.46 | 0.42 |
P-value for heterogeneity.
OR, odds ratio; CI, confidence interval; SCLC, small cell lung cancer; SCC, squamous cell carcinoma; AC, adenocarcinoma.
Figure 2.
Forest plot for the association between the glutathione S-transferase θ1 polymorphism and lung cancer risk in East Asian populations.
Publication bias
As demonstrated in Fig. 3, the shapes of the funnel plots appeared symmetrical in the overall populations, indicating the absence of publication bias. Furthermore, the results of the Egger’s test provided statistical evidence for the funnel plot asymmetry (t=−1.57; P=0.1265).
Figure 3.
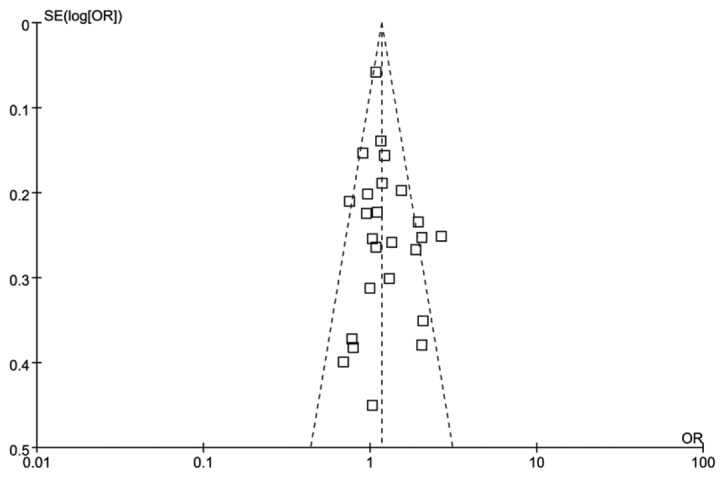
Funnel plot analysis to detect publication bias, indicating the odds ratios for the major effect of glutathione S-transferase θ1.
Discussion
The results of the current meta-analysis indicated that GSTT1 polymorphism was associated with an increased risk of developing lung cancer in the East Asian populations. Furthermore, in subgroups of smoking status, increased lung cancer susceptibility was identified in the smoking population.
The GSTT1 gene is located on chromosome 22q11.2 (47). Individuals carrying homozygous deletions in the GSTT1 genes may present an impaired ability of metabolically eliminating carcinogenic compounds and may, therefore, be at an increased risk of developing lung cancer (6). Since Deakin et al (48) first investigated the GSTT1 polymorphism in lung cancer, a number of studies have been performed to evaluate the association between this homozygous deletion and the risk of developing lung cancer (4,49,50). Certain studies have indicated that this polymorphism in the GSTT1 gene is a risk factor for lung cancer; however, this is inconsistent with the findings of other studies (51–53). Additionally, a previous study demonstrated that individuals harboring the null deletion of the GSTT1 gene had a 2.4-fold higher risk of developing lung cancer (54). Furthermore, in a previous meta-analysis focused on the Asian population, the results were predominantly positive compared with other ethnic groups (55). The disparity in lung cancer susceptibility among different ethnicities with the GSTT1 null genotype is consistent with a number of previous studies indicating that the frequency of the GSTT1 deletion varies among different populations (56,57). In particular, the prevalence of the GSTT1 null genotype is lower among Caucasians (10–20%) compared with Asians (50–60%) (58). Thus, ethnicity may be an important factor influencing the GSTT1 gene sensitivity to lung cancer. To the best of our knowledge, the current study is the first to investigate and determine that the GSTT1 null genotype is a risk factor of lung cancer in the East Asian populations.
Tobacco smoking is known to be one of the major risk factors of lung cancer (56). Tobacco contains a variety of carcinogens, including polycyclic aromatic hydrocarbons, N-nitrosamines and aromatic heterocyclic amines, that are transported through metabolic pathways by the GSTT1 protein (59). A number of studies have identified a higher risk of developing lung cancer in smokers carrying the GSTT1 null genotype (52,60,61). Similarly, the present study identified that the interaction between the GSTT1 gene and tobacco consumption has an effect on the development lung cancer. Furthermore, it has been reported that not all populations were equally susceptible to tobacco-associated carcinogens (62). The frequency of the GSTT1 null genotype was lower in Caucasians compared with Asians, strengthening the hypothesis that polymorphisms in enzymes that metabolize tobacco carcinogens may have a strong association with ethnicity (63).
A number of previous studies have evaluated the effects of gene-gene and gene-environment interactions in lung cancer development. Chen et al (20) indicated that individuals carrying the GSTT1 null genotype combined with other GST mutants may have an enhanced risk of developing lung cancer compared with individuals carrying any mutant alone. For instance, specific studies indicated that a daily diet of fruit and vegetables containing isothiocyanates may reduce the risk of lung cancer among carriers of at least one functional GSTT1 allele (26,64,65). Brennan et al (65) demonstrated that the protective effect of isothiocyanate-containing vegetables was most apparent in individuals with low values of circulating GST enzymes due to the presence of null GSTT1 alleles. In addition, the results are in accordance with various smaller studies of lung cancer, breast cancer and colorectal adenomas that demonstrated a protective effect of isothiocyanates in GSTT1 null carriers (65). More comprehensive studies should be conducted in the future to fully understand the association between cancer risk and isothiocyanate consumption in GSTT1 null individuals.
In the current meta-analysis, various limitations should be acknowledged. Lung cancer is known to be a consequence of multiple risk factors. For instance, lifestyle, diet, age, gender, environment and ethnicity may all contribute as the possible risk factors (48). However, due to the limited participant data provided by the individual studies, the present meta-analysis was not able to conduct a more precise assessment by ruling out those confounding factors. In addition, misclassification of cigarette consumption may have occurred due to the vague definition of cigarette consumption in certain studies. Furthermore, only published studies were included in the current meta-analysis, which may have biased the results.
In conclusion, the present study indicated that the GSTT1 null genotype may contribute towards the increased lung cancer risk in the East Asian populations. Future large and well-designed epidemiological studies considering the potential interactions are required to confirm the results of the present meta-analysis.
References
- 1.Dialyna IA, Miyakis S, Georgatou N, Spandidos DA. Genetic polymorphisms of CYP1A1, GSTM1 and GSTT1 genes and lung cancer risk. Oncol Rep. 2003;10:1829–1835. [PubMed] [Google Scholar]
- 2.Yang X, Qiu MT, Hu JW, et al. GSTT1 null genotype contributes to lung cancer risk in Asian populations: a meta-analysis of 23 studies. PLoS One. 2013;8:e62181. doi: 10.1371/journal.pone.0062181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Litwack G, Ketterer B, Arias IM. Ligandin: a hepatic protein which binds steroids, bilirubin, carcinogens and a number of exogenous organic anions. Nature. 1971;234:466–467. doi: 10.1038/234466a0. [DOI] [PubMed] [Google Scholar]
- 4.Gallegos-Arreola MP, Gómez-Meda BC, Morgan-Villela G, et al. GSTT1 gene deletion is associated with lung cancer in Mexican patients. Dis Markers. 2004;19:259–261. doi: 10.1155/2004/826408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Deng Y, Cheng J, Ding J, Tokudome S. GST genetic polymorphisms and lung adenocarcinoma susceptibility in a Chinese population. Cancer Lett. 2003;201:185–193. doi: 10.1016/S0304-3835(03)00480-4. [DOI] [PubMed] [Google Scholar]
- 6.Salagovic J, Kalina I, Stubna J, et al. Genetic polymorphism of glutathione S-transferases M1 and T1 as a risk factor in lung and bladder cancers. Neoplasma. 1998;45:312–317. [PubMed] [Google Scholar]
- 7.Zhong S, Howie AF, Ketterer B, et al. Glutathione S-transferase mu locus: use of genotyping and phenotyping assays to assess association with lung cancer susceptibility. Carcinogenesis. 1991;12:1533–1537. doi: 10.1093/carcin/12.9.1533. [DOI] [PubMed] [Google Scholar]
- 8.Sprenger R, Schlagenhaufer R, Kerb R, et al. Characterization of the glutathione S-transferase GSTT1 deletion: discrimination of all genotypes by polymerase chain reaction indicates a trimodular genotype-phenotype correlation. Pharmacogenetics. 2000;10:557–565. doi: 10.1097/00008571-200008000-00009. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi S, Watanabe J, Kawajiri K. High susceptibility to lung cancer analyzed in terms of combined genotypes of P450IA1 and Mu-class glutathione S-transferase genes. Jpn J Cancer Res. 1992;83:866–870. doi: 10.1111/j.1349-7006.1992.tb01992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirvonen A, Husgafvel-Pursiainen K, Anttila S, Vainio H. The GSTM1 null genotype as a potential risk modifier for squamous cell carcinoma of the lung. Carcinogenesis. 1993;14:1479–1481. doi: 10.1093/carcin/14.7.1479. [DOI] [PubMed] [Google Scholar]
- 11.Alexandrie AK, Sundberg MI, Seidegård J, et al. Genetic susceptibility to lung cancer with special emphasis on CYP1A1 and GSTM1: a study on host factors in relation to age at onset, gender and histological cancer types. Carcinogenesis. 1994;15:1785–1790. doi: 10.1093/carcin/15.9.1785. [DOI] [PubMed] [Google Scholar]
- 12.Houlston RS. Glutathione S-transferase M1 status and lung cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 1999;8:675–682. [PubMed] [Google Scholar]
- 13.London SJ, Daly AK, Cooper J, et al. Polymorphism of glutathione S-transferase M1 and lung cancer risk among African-Americans and Caucasians in Los Angeles County, California. J Natl Cancer Inst. 1995;87:1246–1253. doi: 10.1093/jnci/87.16.1246. [DOI] [PubMed] [Google Scholar]
- 14.Saarikoski ST, Voho A, Reinikainen M, et al. Combined effect of polymorphic GST genes on individual susceptibility to lung cancer. Int J Cancer. 1998;77:516–521. doi: 10.1002/(SICI)1097-0215(19980812)77:4<516::AID-IJC7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 15.Kiyohara C, Horiuchi T, Takayama K, Nakanishi Y. Genetic polymorphisms involved in carcinogen metabolism and DNA repair and lung cancer risk in a Japanese population. J Thorac Oncol. 2012;7:954–962. doi: 10.1097/JTO.0b013e31824de30f. [DOI] [PubMed] [Google Scholar]
- 16.Tamaki Y, Arai T, Sugimura H, et al. Association between cancer risk and drug-metabolizing enzyme gene (CYP2A6, CYP2A13, CYP4B1, SULT1A1, GSTM1, and GSTT1) polymorphisms in cases of lung cancer in Japan. Drug Metab Pharmacokinet. 2011;26:516–522. doi: 10.2133/dmpk.DMPK-11-RG-046. [DOI] [PubMed] [Google Scholar]
- 17.Piao JM, Shin MH, Kim HN, et al. Glutathione-S-transferase (GSTM1, GSTT1) null phenotypes and risk of lung cancer in a Korean population. Asian Pac J Cancer Prev. 2013;14:7165–7169. doi: 10.7314/APJCP.2013.14.12.7165. [DOI] [PubMed] [Google Scholar]
- 18.Shaikh RS, Amir M, Masood AI, et al. Frequency distribution of GSTM1 and GSTT1 null allele in Pakistani population and risk of disease incidence. Environ Toxicol Pharmacol. 2010;30:76–79. doi: 10.1016/j.etap.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Chan-Yeung M, Tan-Un KC, Ip MS, et al. Lung cancer susceptibility and polymorphisms of glutathione-S-transferase genes in Hong Kong. Lung Cancer. 2004;45:155–160. doi: 10.1016/j.lungcan.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 20.Chen HC, Cao YF, Hu WX, et al. Genetic polymorphisms of phase II metabolic enzymes and lung cancer susceptibility in a population of Central South China. Dis Markers. 2006;22:141–152. doi: 10.1155/2006/436497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiyohara C, Yamamura KI, Nakanishi Y, et al. Polymorphism in GSTM1, GSTT1 and GSTP1 and susceptibility to lung cancer in a Japanese population. Asian Pac J Cancer Prev. 2000;1:293–298. [PubMed] [Google Scholar]
- 22.Lan Q, He X, Costa DJ, et al. Indoor coal combustion emissions, GSTM1 and GSTT1 genotypes and lung cancer risk: a case-control study in Xuan Wei, China. Cancer Epidemiol Biomarkers Prev. 2000;9:605–608. [PubMed] [Google Scholar]
- 23.Lee KM, Kang D, Lee SJ, et al. Interactive effect of genetic polymorphism of glutathione S-transferase M1 and smoking on squamous cell lung cancer risk in Korea. Oncol Rep. 2006;16:1035–1039. [PubMed] [Google Scholar]
- 24.London SJ, Yuan JM, Chung FL, et al. Isothiocyanates, glutathione S-transferase M1 and T1 polymorphisms, and lung-cancer risk: a prospective study of men in Shanghai, China. Lancet. 2000;356:724–729. doi: 10.1016/S0140-6736(00)02631-3. [DOI] [PubMed] [Google Scholar]
- 25.Yang M, Choi Y, Hwangbo B, et al. Combined effects of genetic polymorphisms in six selected genes on lung cancer susceptibility. Lung Cancer. 2007;57:135–142. doi: 10.1016/j.lungcan.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Deng Y, Cheng J, et al. GST genetic polymorphisms and lung adenocarcinoma susceptibility in a Chinese population. Cancer Lett. 2003;201:185–193. doi: 10.1016/S0304-3835(03)00480-4. [DOI] [PubMed] [Google Scholar]
- 27.Wang N, Wu Y, Zhou X, Wu Y. Association between genetic polymorphism of metabolizing enzymes and DNA repairing enzymes and the susceptibility of lung cancer in Henan population. Wei Sheng Yan Jiu. 2012;41:251–256. (In Chinese) [PubMed] [Google Scholar]
- 28.Yuan T, Zhou Q, Zhu W, et al. Relationship between genetic polymorphism of GSTT1 gene and inherent susceptibility to lung cancer in Han population in Sichuan, China. Zhongguo Fei Ai Za Zhi. 2005;8:107–111. doi: 10.3779/j.issn.1009-3419.2005.02.07. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 29.Bai TY. PhD dissertation. Inner Mongolia Medical College; Inner Mongolia: 2011. The study on the polymorphisms of GSTM1, GSTM3, GSTT1, GSTP1 genes and susceptibility to lung cancer in Mongolian population; pp. 15–18. (In Chinese) [Google Scholar]
- 30.Li YF. PhD dissertation. Sichuan University; Sichuan: 2007. A case-control study on the associations between polymorphism of GSTM1, GSTT1 and susceptibility to breast cancer and lung cancer; pp. 26–33. (In Chinese) [Google Scholar]
- 31.Du GB. PhD dissertation. North Sichuan Medical University; Sichuan: 2011. A study on relationship between genetic polymorphism of GSTM1 and GSTT1 gene and susceptibility to lung cancer in the population of northern Sichuan of China; pp. 18–25. (In Chinese) [Google Scholar]
- 32.Wang N. PhD dissertation. Zhengzhou University; Zhengzhou: 2003. The relationship between the deletion of GSTM1, GSTT1 and susceptibility to lung cancer; pp. 18–26. (In Chinese) [Google Scholar]
- 33.Fan J. PhD dissertation. GuangXi Medical University; GuangXi: 2011. Study on the polymorphisms of GSTM1 and GSTT1 genes associated with susceptibility to lung cancer; pp. 15–24. (In Chinese) [Google Scholar]
- 34.Liu JN. PhD dissertation. Yanbian University; Yanbian: 2009. Study on the relationship between the genetic polymorphism of GSTT1 gene, smoking and lung cancer susceptibility; pp. 8–12. (In Chinese) [Google Scholar]
- 35.He DX, Tang Y. The relationship of GSTT1 polymorphism and chromosome 15 aberration in lung cancer patients. Zhong Liu Fang Zhi Yan Jiu. 2006;33:308–310. (In Chinese) [Google Scholar]
- 36.Qi XS, Lv HM, Xia Y, et al. A primary case-control study on the relationship between genetic polymorphisms of GSTT1 and lung cancer susceptibility to the people living in high radon-exposed area. Zhong Guo Zhi Ye Yi Xue. 2008;31:361–367. (In Chinese) [Google Scholar]
- 37.Lan Q, He XZ, Costa D, et al. Glutathione S-transferase GSTM1 and GSTT1 genotypes and susceptibility to lung cancer. Wei Sheng Yan Jiu. 1999;28:9–11. [PubMed] [Google Scholar]
- 38.Zhang JK. PhD dissertation. Jinan University; Guangzhou: 2002. Genetic polymorphisms of glutathione S-transferase M1 and T1 gene related with the susceptibility to lung cancer; pp. 25–32. (In Chinese) [Google Scholar]
- 39.Fan J, Gan LZ, Liang KC, Liang XM. Relationship of GSTM1 and GSTT1 genetic polymorphisms with lung cancer susceptibility in Guangxi Zhuang population. Zhong Liu Xue Za Zhi. 2010;16:922–925. (In Chinese) [Google Scholar]
- 40.Cao YF, Chen HC, Liu XF, et al. Study on the relationship between the genetic polymorphisma of GSTM1 and GSTT1 genes and lung cancer susceptibility in the population of Hunan province of China. Life Sci Res. 2004;8:126–132. (In Chinese) [Google Scholar]
- 41.Liang KC, Gan LK, Ruan L, et al. Correlational research of the relationship between the genetic polymorphism of GSTM1 and GSTT1 in the Zhuang population and lung cancer. Acta Med Sinica. 2012;25:813–817. (In Chinese) [Google Scholar]
- 42.Wells GA, Shea B, O’Connell D, et al. Ottawa Hospital Research Institute. [Accessed Oct 19, 2009];The Newcastle-Ottawa Scale (NOS) for assessing the quality if nonrandomized studies in meta-analyses. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
- 43.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 44.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 45.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 46.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rebbeck TR. Molecular epidemiology of the human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1997;6:733–743. [PubMed] [Google Scholar]
- 48.Deakin M, Elder J, Hendrickse C, et al. Glutathione S-transferase GSTT1 genotypes and susceptibility to cancer: studies of interactions with GSTM1 in lung, oral, gastric and colorectal cancers. Carcinogenesis. 1996;17:881–884. doi: 10.1093/carcin/17.4.881. [DOI] [PubMed] [Google Scholar]
- 49.Sunaga N, Kohno T, Yanagitani N, et al. Contribution of the NQO1 and GSTT1 polymorphisms to lung adenocarcinoma susceptibility. Cancer Epidemiol Biomarkers Prev. 2002;11:730–738. [PubMed] [Google Scholar]
- 50.Harms C, Salama SA, Sierra-Torres CH, et al. Polymorphisms in DNA repair genes, chromosome aberrations and lung cancer. Environ Mol Mutagen. 2004;44:74–82. doi: 10.1002/em.20031. [DOI] [PubMed] [Google Scholar]
- 51.Stücker I, Hirvonen A, de Waziers I, et al. Genetic polymorphisms of glutathione S-transferases as modulators of lung cancer susceptibility. Carcinogenesis. 2002;23:1475–1481. doi: 10.1093/carcin/23.9.1475. [DOI] [PubMed] [Google Scholar]
- 52.Yang P, Bamlet WR, Ebbert JO, et al. Glutathione pathway genes and lung cancer risk in young and old populations. Carcinogenesis. 2004;25:1935–1944. doi: 10.1093/carcin/bgh203. [DOI] [PubMed] [Google Scholar]
- 53.Cabral RE, Caldeira-de-Araujo A, Cabral-Neto JB, Costa Carvalho Mda G. Analysis of GSTM1 and GSTT1 polymorphisms in circulating plasma DNA of lung cancer patients. Mol Cell Biochem. 2010;338:263–269. doi: 10.1007/s11010-009-0360-6. [DOI] [PubMed] [Google Scholar]
- 54.Sreeja L, Syamala V, Hariharan S, et al. Possible risk modification by CYP1A1, GSTM1 and GSTT1 gene polymorphisms in lung cancer susceptibility in a South Indian population. J Hum Genet. 2005;50:618–627. doi: 10.1007/s10038-005-0303-3. [DOI] [PubMed] [Google Scholar]
- 55.Langevin SM, Ioannidis JP, Vineis P, Taioli E Genetic Susceptibility to Environmental Carcinogens group (GSEC) Assessment of cumulative evidence for the association between glutathione S-transferase polymorphisms and lung cancer: application of the Venice interim guidelines. Pharmacogenet Genomics. 2010;20:586–597. doi: 10.1097/FPC.0b013e32833c3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raimondi S, Paracchini V, Autrup H, Barros-Dios JM, Benhamou S, Boffetta P, et al. Meta- and pooled analysis of GSTT1 and lung cancer: a HuGE-GSEC review. Am J Epidemiol. 2006;164:1027–1042. doi: 10.1093/aje/kwj321. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Yang H, Li L, Wang H. Glutathione S-transferase T1 gene deletion polymorphism and lung cancer risk in Chinese population: a meta-analysis. Cancer Epidemiol. 2010;34:593–597. doi: 10.1016/j.canep.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 58.Nelson HH, Wiencke JK, Christiani DC, et al. Ethnic differences in the prevalence of the homozygous deleted genotype of glutathione S-transferase theta. Carcinogenesis. 1995;16:1243–1245. doi: 10.1093/carcin/16.5.1243. [DOI] [PubMed] [Google Scholar]
- 59.Zhong S, Howie AF, Ketterer B, et al. Glutathione S-transferase mu locus: use of genotyping and phenotyping assays to assess association with lung cancer susceptibility. Carcinogenesis. 1991;12:1533–1537. doi: 10.1093/carcin/12.9.1533. [DOI] [PubMed] [Google Scholar]
- 60.Spitz MR, Duphorne CM, Detry MA, et al. Dietary intake of isothiocyanates: evidence of a joint effect with glutathione S-transferase polymorphisms in lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2000;9:1017–1020. [PubMed] [Google Scholar]
- 61.Hou SM, Fält S, Nyberg F. Glutathione S-transferase T1-null genotype interacts synergistically with heavy smoking on lung cancer risk. Environ Mol Mutagen. 2001;38:83–86. doi: 10.1002/em.1054. [DOI] [PubMed] [Google Scholar]
- 62.Spitz MR, Wei Q, Dong Q, et al. Genetic susceptibility to lung cancer: the role of DNA damage and repair. Cancer Epidemiol Biomarkers Prev. 2003;12:689–698. [PubMed] [Google Scholar]
- 63.Honma HN, De Capitani EM, Perroud MW, et al. Influence of p53 codon 72 exon 4, GSTM1, GSTT1 and GSTP1*B polymorphisms in lung cancer risk in a Brazilian population. Lung Cancer. 2008;61:152–162. doi: 10.1016/j.lungcan.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 64.Zhao B, Seow A, Lee EJ, Poh WT, Teh M, Eng P, et al. Dietary isothiocyanates, glutathione S-transferase -M1, -T1 polymorphisms and lung cancer risk among Chinese women in Singapore. Cancer Epidemiol Biomarkers Prev. 2001;10:1063–1067. [PubMed] [Google Scholar]
- 65.Brennan P, Hsu CC, Moullan N, Szeszenia-Dabrowska N, Lissowska J, Zaridze D, Jr, et al. Effect of cruciferous vegetables on lung cancer in patients stratified by genetic status: a mendelian randomisation approach. Lancet. 2005;366:1558–1560. doi: 10.1016/S0140-6736(05)67628-3. [DOI] [PubMed] [Google Scholar]