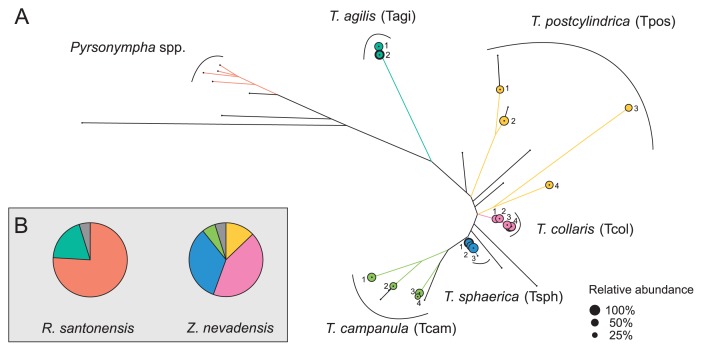
Fig. 4.
(A) Maximum-likelihood tree of all Endomicrobia phylotypes (99% sequence similarity) detected in whole-genome amplification (MDA) products of all single flagellate cells. Branches shared by phylotypes from the same host species are color coded; the area of the circles indicates the relative abundance of the major phylotype in each library. Black dots indicate the position of phylotypes that originated from whole-gut samples of Zootermopsis nevadensis or Reticulitermes santonensis. (B) Pie charts indicate the relative abundance of the different Endomicrobia clusters in the whole gut samples. The colors represent the major phylotypes from selected Trichonympha cells; phylotypes from whole-gut samples are shown in grey. Phylotypes in the Pyrsonympha cluster were identified based on the sequences retrieved from selected flagellates from R. santonensis (unpublished results).