Fig. 2.
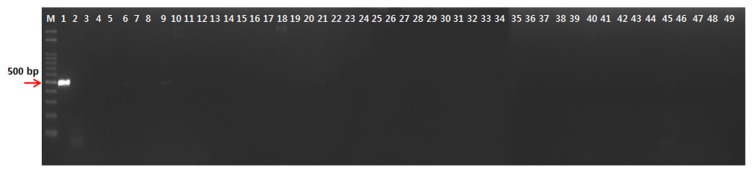
Gel electrophoresis of the polymerase cain reaction products formed with primer Pc-12-F/Pc-12-R and total DNA of lane 1, Pseudomonas coronafaciens LMG 5060, lanes 2–31, P. syringae pvs, actinidiae KACC 10582, antirrhini ICMP 4303, aptata DSM 50252, atrofaciens ICMP 4394, berberidis NCPPB 2724, ciccaronei NCPPB 2355, delphhinii ICMP 529, dysoxyli ICMP 545, eriobotyae NCPPB 2331, helianthi NCPPB 1229, japonica ICMP 6305, lachrymans ATCC 11965, lapsa ATCC 10859, maculicola ICMP 3935, mellea ICMP 5711, mori ICMP 4331, morsprunorum ICMP 5795, myricae ICMP 7118, panici NCPPB 1498, papulans ICMP 4040, passiflorae NCPPB 1386, persicae NCPPB 2761, pisi ICMP 4433, ribicola NCPPB 963, sesami NCPPB 1016, syringae NCPPB 388, tabaci ICMP 2835, tagetis ICMP 4091, tomato NCPPB 2683, ulmi NCPPB 632; lanes 32~34, P. savastanoi pvs. glycinea NCPPB 1134, pahseolicola KACC 10575, and savastanoi NCPPB 639; lane 35, Acidovorax avenae subsp. avenae NCPPB 1011; lanes 36–38, Clavibacter michiganensis subsp. insidiosus NCPPB 1020, michiganensis NCPPB 1064, sepedonicus NCPPB 2137; lane 39, Pectobacterium carotovorum subsp. carotovorum, NCPPB 312; lanes 40–43, Rhizobium radiobacter DSM 30205, R. rhizogenes ATCC 11325, R. rubi NCPPB 1854, R. vitis NCPPB 3554; lane 44, Rhodococcus fascians LMG 3601; lane 45, Ralstonia solanacearum NCPPB 339; lanes 46–47, Xanthomonas campestris pvs. campestris KACC 10377, vesicatoria KACC 11157; lane 48, X. oryzae pv. oryzae KACC 10331; lane 1, P. coronafaciens LMG 5060, as a positive control; lane 49, water as a negative control.
