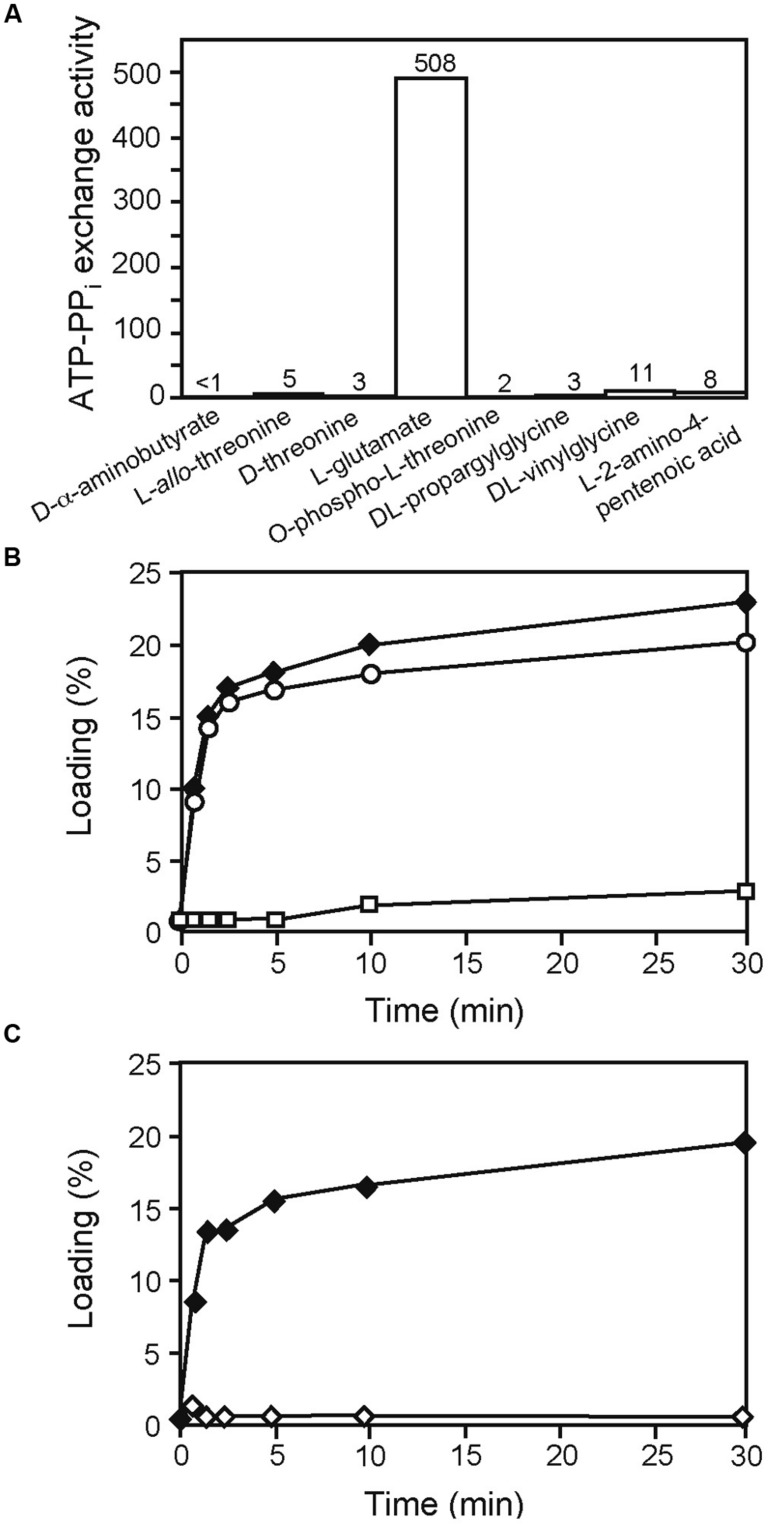
FIGURE 4.
Identification of L-Glu as the amino acid substrate activated by AmbE and loaded onto its T1 domain. (A) End-point ATP-[32P] pyrophosphate exchange assay to screen potential AmbE substrates. At 120 min, generated ATP (formed by the reverse reaction of the A domain of AmbE in the presence of [32P] pyrophosphate) was bound and counted. ATP-PPi exchange activity is expressed as the amount of conversion (one being 100% conversion) multiplied by the molar ratio of PPi to AmbE. (B) Loading of 14C-L-Glu onto AmbE (filled diamonds), AmbES1286A (empty rectangles), and AmbES1819A (empty circles). Proteins were incubated with the radiolabeled amino acid in an aminoacylation assay. At several time points, proteins (with bound amino acids) were precipitated and radioactivity was counted. Percentage of loading was determined using the molar ratio of bound radioactivity to the amount of protein in the assay. (C) Mutations in the A domain of AmbE abolish protein loading with L-Glu. AmbES1958A (filled diamonds), and AmbED644A,K1230T,S1958A (empty diamonds) were incubated with 14C-L-Glu in an aminoacylation assay. At several time points, proteins (with bound amino acids) were precipitated and radioactivity was counted. Percentage of loading was determined using the molar ratio of bound radioactivity to the amount of protein in the assay. Note that the mutation of the active site Ser (S1958) of the TE domain does not interfere with loading of L-Glu. By contrast, amino acid alterations in the A domain (D644A and K1230T) abolished loading entirely.