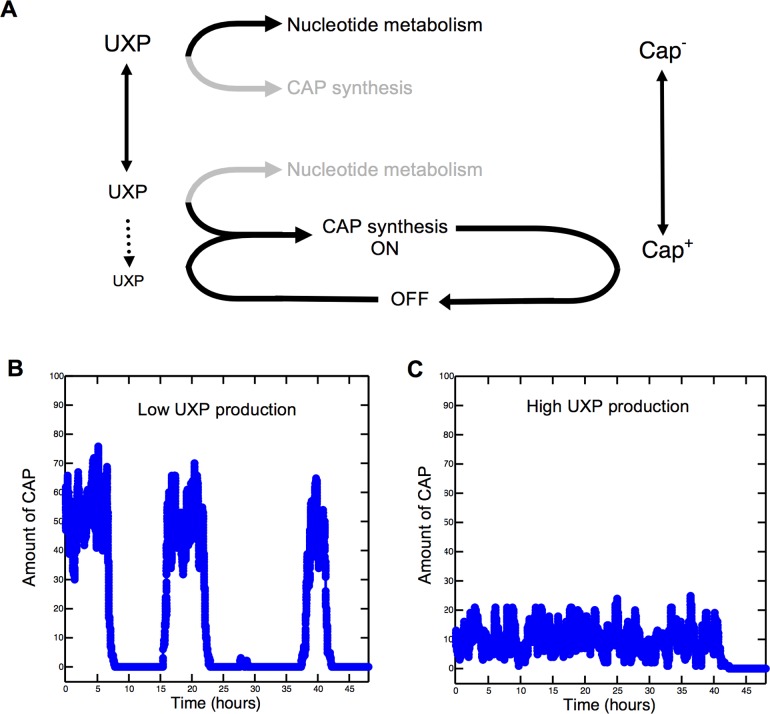
Fig 6. The growth-capsulation model for stochastic capsule switching.
(A) A schematic model for capsule switching. Central to the switch is the bifurcation of the UTP pool into nucleotide metabolism (via PyrG) and CAP synthesis (via GalU). When UXP (a signal molecule proportional to UTP) levels are high, cells commit resources to nucleotide metabolism, and expression of the GalU branch is basal. As UXP levels fall, the likelihood that CAP synthesis is activated increases because of the accumulation of a positive regulator sensitive to UXP levels. Once a cell commits to CAP synthesis, nucleotide metabolism slows, and continued CAP synthesis positively feeds back to maintain low UXP levels. To enable a return to Cap-, we model negative feedback via a repressor activated concomitantly with CAP synthesis. The length of time cells remain Cap+ depends on many factors that are each subject to molecular noise. This introduces stochasticity to the system; we model this using the Gillespie algorithm (see Results text for more detail). (B) A simulation of the model showing that low UXP production can induce switching through changes in CAP expression. Low UXP promotes CAP expression through increased availability of a presumed transcriptional activator of CAP biosynthetic genes. Cap+ persists until the CAP repressor, induced by CAP, is synthesized and outcompetes the activator. Bound repressor causes CAP mRNA levels to fall because of halted synthesis and degradation. Cap- persists until repressor concentration decreases and the DNA binding site is free. UXP is low, and the cycle repeats. See also S5 Fig, S1 Code, and S2 Code. (C) Same as B but with elevated UXP production. The positive regulator-repressor system is not utilized; thus, CAP expression remains basal.