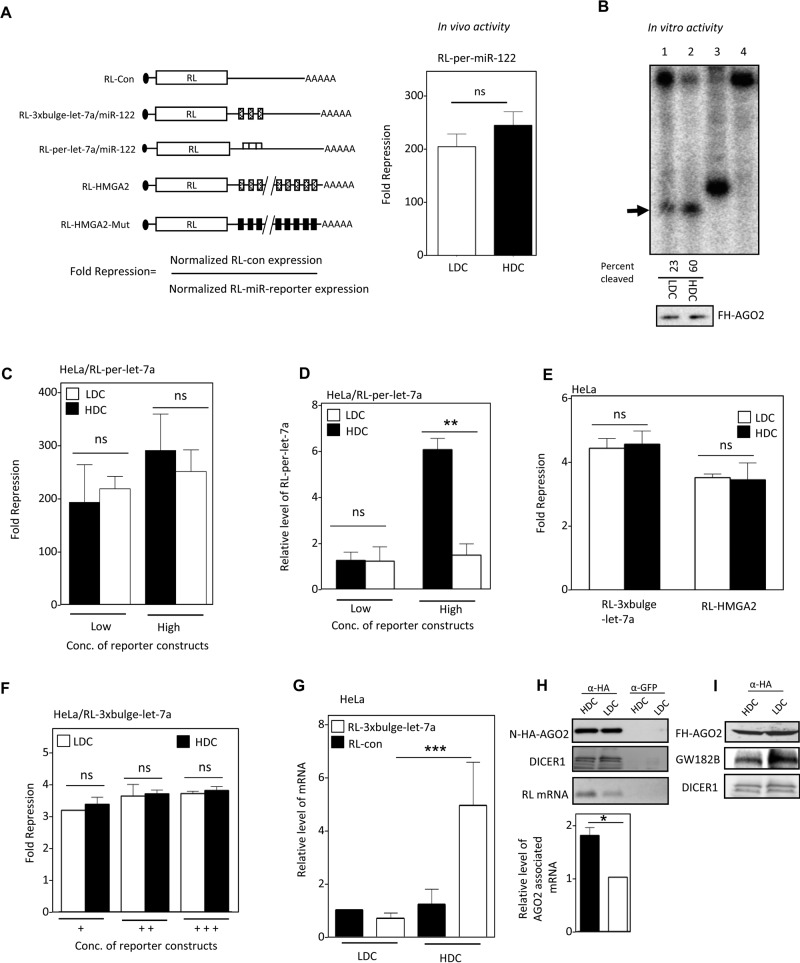
FIGURE 2:
Defective miRNA-mediated repression in HDC human cells. (A) Reporter mRNAs used to measure miRNA activity, showing how fold repression was measured (left). In vivo RISC activity of miR-122 in HDC and LDC HeLa cells expressing miR-122 and an RL reporter with one perfect miR-122 site (right). (B) RISC cleavage activity of miRISC-122 isolated from LDC and HDC HeLa cells cotransfected with FH-AGO2 construct and miR-122 expression plasmid. The activities were measured and quantified in an in vitro RISC cleavage reaction using 5′-32P-labeled miR-122 target RNA. Relative quantification of the RISC activity isolated from LDC and HDC HeLa cells (lanes 1 and 2) was done by densitometric analysis. Lane 4, no RISC; in lane 3, a 21-nt-long radiolabeled DNA oligo was used as size marker. The RISC-cleaved band is marked by an arrowhead. Western blot for the isolated RISC with anti-HA antibody confirmed equal levels of FH-AGO2 protein in each reaction (1 and 2). (C) Invariant repression levels of let-7a miRNA reporter in HDC and LDC cells. Cells transfected with low and high concentrations of plasmids encoding the RL reporter with one perfect let-7a binding site were grown to HDC and LDC, and the normalized repression values were measured in HeLa cells. (D) Reduced cleavage of mRNA targets with perfect miRNA site in HDC condition. Real-time quantification of RL mRNA level was performed in RNA isolated from RL-per-let-7a–transfected HeLa cells expressing low and high concentrations of reporter and grown to HDC and LDC conditions. The 18S rRNA normalized relative levels of RL mRNA for individual samples are plotted. Values in the LDC state are set to 1. (E) Relative repression level of two let-7a reporters with multiple imperfect let-7a binding sites in HeLa cells from HDC and LDC conditions. RL-HMGA2 has a 3′ UTR of let-7a target gene HMGA2. RL-HMGA2-Mut with mutated let-7a sites served as control. For RL-3xbulge-let-7a, RL-Con was used as control. (F) Relative fold repression of RL reporter with three imperfect let-7a binding sites in cells in which an increasing amount of reporter-encoding plasmid was used for transfection of HeLa cells. (G) Real-time PCR detection of RL mRNA in RL-3xbulge-let-7a– or RL-Con–transfected HeLa cells under similar experimental conditions as in E. (H) Association of target mRNA with AGO2 in HDC and LDC HeLa cells expressing RL-3xbulge-let-7a. N-HA-AGO2 was immunoprecipitated with anti-HA antibody from cells coexpressing it along with RL reporter. Control immunoprecipitation reaction was performed with anti-GFP antibody. Western blot analysis of the IPed AGO2 was performed, and associated DICER1 was detected (top). RNA extracted from the lysates was used to determine the amount of RL mRNA associated with AGO2 by PCR (top) and also quantified by real-time PCR (bottom). (I) Association of GW182B with AGO2 in HDC and LDC cells. Cells expressing FH-AGO2 were grown to HDC and LDC states, HA-AGO2 was immunoprecipitated with anti-FLAG antibody, and associated GW182B was detected. All experiments were performed a minimum of three times. Here Low and High refer, respectively, to 25 and 100 ng of plasmids used for transfection of cells present per well of a six-well plate. In F, +, ++, and +++ represent respectively 25, 100, and 500 ng of plasmid transfected per well of a six-well plate. For all relevant panels, ns, nonsignificant, *p < 0.05, **p < 0.01, and ***p < 0.0001. The p values were determined by paired t test, and results depict mean values from at least three experiments.