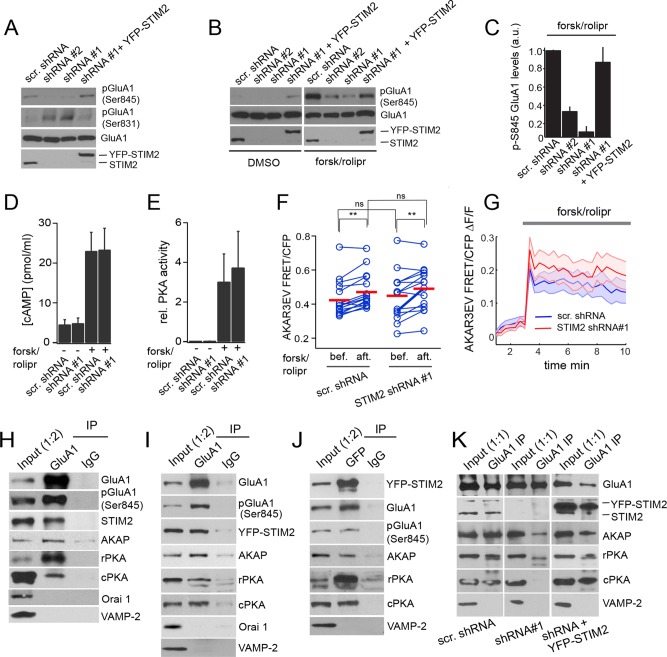
FIGURE 4:
Reciprocal regulation of GluA1 Ser-831 and Ser-845 phosphorylation by STIM2. (A–C) Immunoblot analysis of hippocampal neurons (DIV 21) transduced with the indicated shRNAs and YFP-STIM2 for rescue experiments. (A) Decreased pSer-845 and increased pSer-831 in neurons expressing STIM2 shRNA#1 and #2. Both phosphorylation phenotypes are rescued by coexpression of YFP-STIM2. (B) Immunoblot analysis of GluA1 pSer-845 in cells treated with DMSO (vehicle) or 50 μM forskolin/0.1 μM rolipram for 30 min (same blot exposure for all conditions). (C) Quantification of GluA1 phospho–Ser-845 signals by densitometry after forsk/rolipr treatment (n = 4). (D, E) cAMP levels and PKA activity measured in DMSO or forsk/rolipr-treated neurons (DIV 20–21) transduced with the indicated shRNAs (n = 5 for each condition). (F, G) AKAR3EV FRET measurements in neurons (DIV 21) electroporated with the indicated shRNA constructs. (F) Pairwise analysis of AKAR3EV FRET in individual neurons before and 3 min after forsk/rolipr addition in control (n = 17) and STIM2-silenced cells (n = 15). Red bars indicate the mean. Cells were analyzed from three independent experiments. **p < 0.01, paired t tests. ns, nonsignificant (p > 0.05). (G) Fold change in AKAR3EV FRET induced by forsk/rolipr. The shaded areas represent SEM. The average fold increase in FRET in control and STIM2-silenced cells is not statistically different, p > 0.05, t test. (H–K) Co-IPs from adult rat brains (H) or DIV 21 hippocampal neurons (I–K) transduced with YFP-STIM2 (I, J) and the indicated shRNAs (K). (H, I, K) IPs with a GluA1 Ab or a control immunoglobulin G (IgG). (J) IP using anti-GFP Ab or a control IgG. Fractions were immunoblotted with the indicated Abs. The ratio indicated in the input lane reflects the fraction of input loaded relative to the IP fraction. See also Supplemental Figures S3 and S4 and Supplemental Movies S1 and S2.