Figure 3.
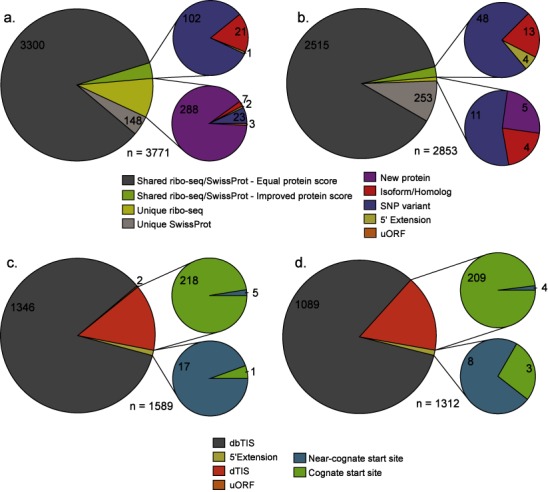
PROTEOFORMER enables deep proteome coverage. Pie charts representing the number of protein and peptide identifications obtained from the shotgun proteomics and N-terminal COFRADIC experiments based on searching the PROTEOFORMER + Swiss-Prot database for both mouse ESC cells and human HCT116 cells using a 1% FDR threshold. Execution times of the different modules used in order to arrive at these results can be found in Supplementary Table S3. (a) Shotgun proteomics results (mouse). A total of 3771 proteins were identified. (b) Shotgun proteomics results (human), identifying a total of 2853 proteins. For the shotgun experiments, a categorization was made based on the fact that the protein can be picked up using the PROTEOFORMER and/or Swiss-Prot sequence database. Also, the improved and new protein identifications were further classified into the following categories: new, isoform/homolog, SNP variant, 5′ extension and uORF. (c) N-terminal COFRADIC (mouse) experiment resulting in 1589 N-terminal peptide identifications. (d) N-terminal COFRADIC results (human). Here, 1312 N-termini were identified. The N-termini were categorized as either dbTIS (database annotated TIS), dTIS (downstream TIS), 5′ extension or uORF.
