Figure 4.
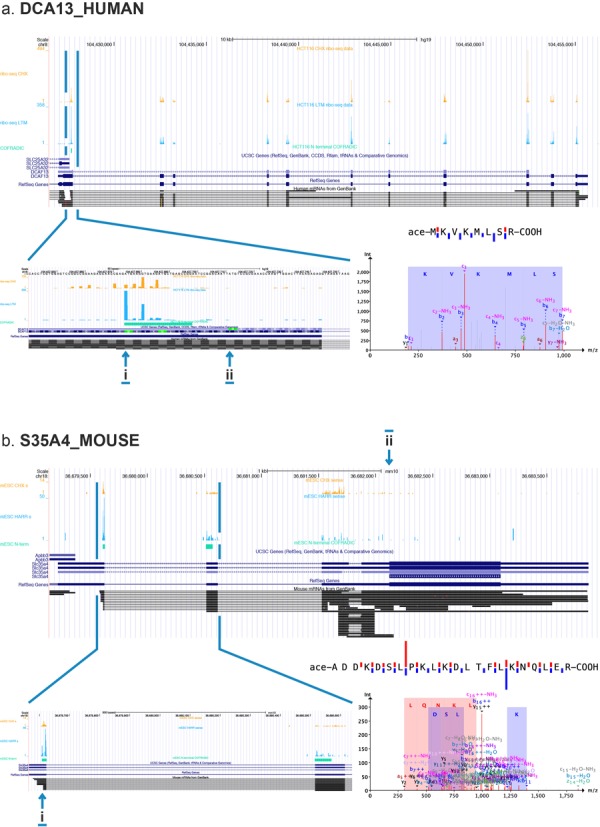
Examples of new proteoforms. (a) N-terminal extension of DCA13_HUMAN and (b) translated uORF of S35A4_MOUSE that were picked up by the proteogenomics analysis and validated by N-terminal COFRADIC. The UCSC genome browser was used to create a view of the RIBO-seq and COFRADIC data. The different tracks are from top to bottom: CHX-treated RIBO-seq data, LTM/HARR-treated RIBO-seq data, N-terminal COFRADIC data, UCSC genes, RefSeq genes and mRNA. The zoomed-in images show the alternative start site (i, alternative start site; ii, canonical start site), while the MS/MS spectra and sequence fragmentation plots display the confidence and quality of the N-terminal peptide identifications.
