Figure 2.
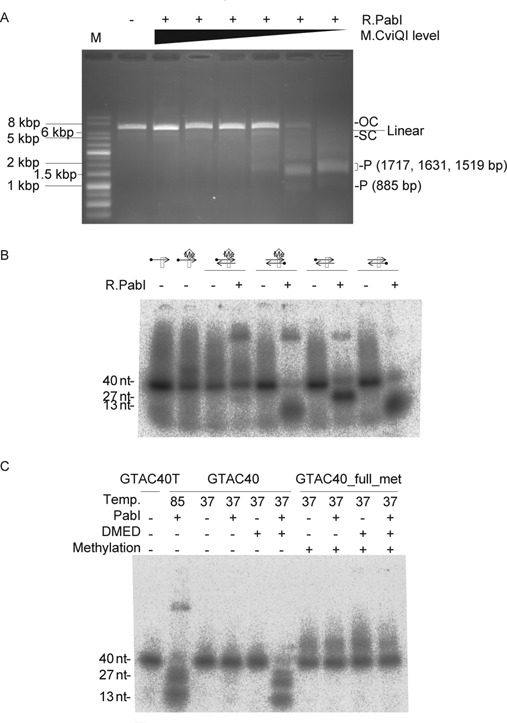
Inhibition of R.PabI activities by methylation. (A) Inhibition of strand cleavage. Plasmid pBAD30_cviQIM (Supplementary Figure S1) with a gene for M.CviQI, which generates 5′-GTm6AC as M.PabI does, under control of the pBAD promoter was prepared from cultures with varying concentrations of arabinose, its inducer. After incubation with R.PabI at 85°C for 6 h, the plasmid DNAs were subjected to 0.8% agarose gel electrophoresis. OC, open circle; SC, supercoiled; P, product DNA. Left lane: 1 kb DNA Ladder. (B) Strand-specific inhibition of cleavage in hemimethylated double-stranded DNA. A 40-mer single-stranded (GTAC40T or GTAC40Tme, Supplementary Table S2) or double-stranded (GTAC40_hemi_met or GTAC40, Supplementary Table S2) substrate (1 pmol, 100 nM) with a 32P-label (black dot) at the 5′-end of either strand was incubated with R.PabI (9.2 pmol, 920 nM) at 85°C for 3 h. Products were separated by 10% denaturing PAGE. Box, recognition sequence (5′-GTAC). Me diamond, methylation of the top strand. Cleavage at the recognition sequence resulted in 27-mer and 13-mer oligonucleotides. The supershifted bands near the top of the gel are likely DNA–R.PabI complexes (see also Figure 5 and related text). (C) Inhibition of DNA glycosylase. A 40-mer double-stranded substrate (GTAC40 or GTAC40_full_met (Supplementary Table S2), 1 pmol, 100 nM) with a 32P -label at the 5′ end of both strands was incubated with R.PabI (9.2 pmol, 920 nM) and then treated with 0.1 M DMED at 37°C for 1 h. Products were separated by 10% denaturing PAGE.
