Figure 4.
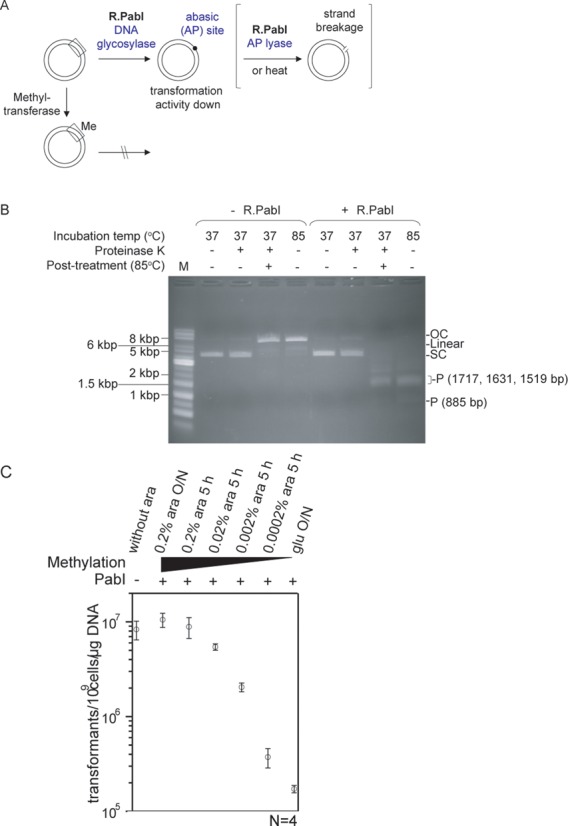
R.PabI restricted DNA without strand breaks. (A) Design. Double-stranded, circular plasmid DNA was treated with R.PabI and transformation activity is measured. Box, recognition sequence. Me, base methylation within the recognition sequence. Only one recognition sequence is shown for simplicity. (B) R.PabI treatment at 37°C does not cause single-strand breaks. Plasmid pBAD30_cviQIM (0.11 pmol, 11 nM, Supplementary Figure S1) purified from E. coli under noninducing conditions was incubated with or without R.PabI (0.77 pmol, 77 nM) at 37°C for 30 min, which was followed by proteinase K (ProK) treatment at 37°C overnight to inactivate R.PabI. DNA was incubated at 85°C for 6 h. Alternatively, plasmid was incubated with or without R.PabI at 85°C for 6 h. Samples were subjected to 0.8% agarose gel electrophoresis. OC, open circle; SC, supercoiled circle; P, product DNA. Left lane: 1 kb DNA ladder. (C) Loss of DNA transformation after R.PabI treatment. pBAD30_cviQIM (0.11 pmol, 11 nM, Supplementary Figure S1) with varying levels of methylation was treated with R.PabI (0.77 pmol, 77 nM) at 37°C for 30 min and purified for quantitative transformation (‘Materials and Methods’ section, Supplementary Figure S5) of E. coli. The plasmid was prepared from cultures with 0.2% arabinose overnight, 0.2% arabinose 5 h, 0.02% arabinose 5 h, 0.002% arabinose 5 h, 0.0002% arabinose 5 h, and glucose overnight, in the order of expected decreased methylation level. E. coli HST08 was transformed with R.PabI-treated plasmids, and resulting colonies were counted to determine transformation efficiencies.
