Figure 1.
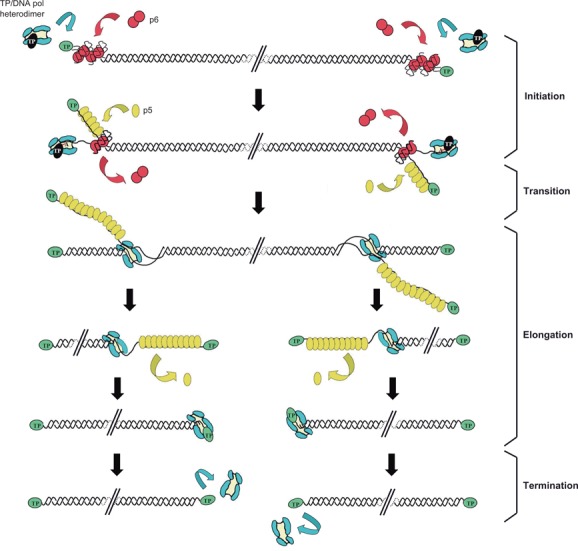
Schematic representation of the mechanism of ϕ29 DNA replication in vitro. The primer TP/DNA polymerase heterodimer recognises the p6-complexed replication origins. The DNA polymerase then catalyses the covalent linkage between dAMP and the hydroxyl group of TP Ser232. After a transition step (not drawn in the figure), the DNA polymerase dissociates from the TP and continues processive elongation coupled to strand displacement. Viral protein SSB p5 binds to the ssDNA-displaced strands and is further removed during the polymerisation process. Continuous elongation by the DNA polymerase from both ends leads to the complete duplication of parental strands. Green ovals: parental TP; black ovals: primer TP; red circles: p6; blue: DNA polymerase; yellow ovals: SSB p5. Linear dsDNA is shown as a double helix.
