Figure 1.
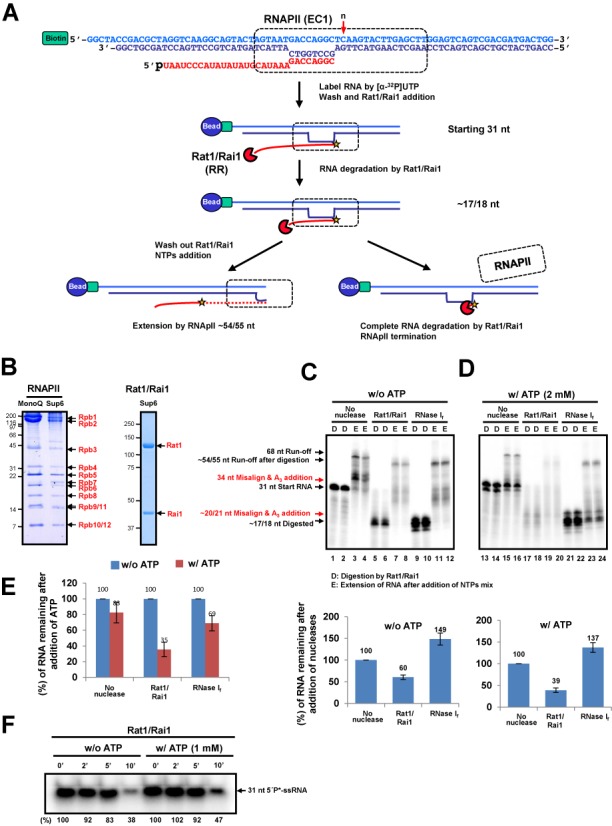
Rat1/Rai1 terminates RNAPII in vitro in the presence of ATP. (A) In vitro transcription termination assay scheme. The EC was assembled with double-stranded DNA (EC1), 5′ phosphorylated RNA and purified RNAPII and was subsequently coupled to magnetic beads. The 3′-end of RNA was radioactively labeled by RNAPII. Rat1/Rai1 digested RNA from 5′ to 3′ up to the surface of RNAPII. If Rat1/Rai1 failed to terminate RNAPII, the polymerase would elongate further using NTPs. However, if Rat1/Rai1 terminated RNAPII, the remaining RNA, which was protected by RNAPII, would be completely degraded by Rat1/Rai1. The red arrow specifies the position (n) of the first incoming NTP. (B) Purified RNAPII complex and Rat1/Rai1 on gels stained with Coomassie. (C) Representative gel image of the in vitro transcription termination assay with Rat1/Rai1 treatment in the absence of ATP. Rat1/Rai1 did not terminate RNAPII by itself. Black arrows indicate the RNAs predicted in (A). Red arrows show ATP-misincorporated RNAs. (D) digestion by Rat1/Rai1. (E) Extension of RNA after addition of NTPs mixture. Quantification of the remaining RNA compared with the control lacking nuclease (set to 100%) is shown below. (D) Rat1/Rai1 terminates RNAPII efficiently in the presence of 2 mM ATP. (E) Quantification of the remaining RNAs after ATP treatment compared with the no ATP control (set to 100%). (F) Time course showing the degradation of 31-nt single strand RNA substrate by Rat1/Rai1 in the absence or presence of 1 mM ATP. The amount of remaining RNA is presented as a percentage of that from the 0 min reaction after Rat1/Rai1 treatment. Asterisk represents radioactive labeling at 5′end of 31-nt RNA.
